Abstract
The role of general transcription factor TFIIB in transcription extends well beyond its evolutionarily conserved function in initiation. Chromatin localization studies demonstrating binding of TFIIB to both the 5’ and 3’ ends of genes in a diverse set of eukaryotes strongly suggested a rather unexpected role of the factor in termination. TFIIB indeed plays a role in termination of transcription. TFIIB occupancy of the 3’ end is possibly due to its interaction with the termination factors residing there. Interaction of the promoter-bound TFIIB with factors occupying the 3’ end of a gene may be the basis of transcription-dependent gene looping. The proximity of the terminator-bound factors with the promoter in a gene loop has the potential to terminate promoter-initiated upstream anti-sense transcription thereby conferring promoter directionality. TFIIB, therefore, is emerging as a factor with pleiotropic roles in the transcription cycle. This could be the reason for preferential targeting of TFIIB by viruses. Further studies are needed to understand the critical role of TFIIB in viral pathogenesis in the context of its newly identified roles in termination, gene looping and promoter directionality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription is an essential step in expression of genetic information in living organisms and is also a process often altered in a plethora of diseases including cancer and virus-related ailments (Liu et al. 2020). An in-depth functional analyses of factors that regulate transcription is crucial to understanding the molecular basis underlying diseases, development and homeostasis. The focus of this review is TFIIB, which is a general transcription factor of RNA polymerase II (RNAPII) transcription machinery. Recent studies revealing novel roles of TFIIB in transcription and its involvement in viral infection have the potential to change the way we currently think about transcription as a process regulated by factors with rigid, step-specific roles.
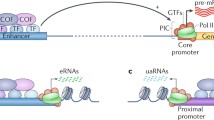
In eukaryotes, RNAPII transcribes DNA to mRNA. Transcription by RNAPII is comprised of a number of steps (Fig. 1). First, is the recruitment of RNAPII to the DNA, made possible by a suite of general transcription factors (GTFs). RNAPII along with the GTFs constitute the preinitiation complex (PIC). The GTFs, in order of recruitment to the promoter, are TFIID, TFIIA, TFIIB, TFIIF, TFIIE and TFIIH (Woychik and Hampsey 2002; Krishnamurthy and Hampsey 2009; Luse 2014).
Following the assembly of the PIC, initiation of transcription takes place. RNAPII must be released from the initiation complex in a coordinated event known as promoter clearance, leaving behind some of the GTFs on the promoter as a ‘scaffold’ (Yudkovsky et al. 2000; Hahn and Young 2011). Following promoter clearance, RNAPII enters the elongation phase. Elongation is marked by RNAPII synthesizing a growing RNA chain as it proceeds along the body of the gene toward the 3’ end (Jonkers and Lis 2015). When the polymerase reaches the terminator region at the 3’ end of a gene, the process of termination takes place. Multi-subunit protein complexes such as the cleavage and polyadenylation factor (CPF) complex, cleavage factor I (CFI) complex, and ribonucleic acid trafficking 1 (Rat1) complex facilitate termination in budding yeast (Mischo and Proudfoot 2013; Baejen et al. 2017). There are orthologous termination complexes in higher eukaryotes (Richard and Manley 2009; Kuehner et al. 2011). Proper termination is vital to the production and nucleocytoplasmic transport of mature mRNA transcripts (Hammell et al. 2002; Mapendano et al. 2010; Lykke-Andersen et al. 2011; Al-Husini et al. 2020).
The focus of this review is the general transcription factor TFIIB, which in addition to its well-established role in initiation, has been implicated in multiple aspects of the transcription cycle (Wang et al. 2010; Medler et al. 2011; Henriques et al. 2012; Allepuz-Fuster et al. 2019). Furthermore, TFIIB has also been found crucial for viral pathogenesis (O’Brien and Ansari, 2021). It is critical to understand the non-canonical roles of TFIIB to fully comprehend the process of transcription, and the molecular basis underlying the role of TFIIB in viral pathogenesis.
TFIIB
TFIIB is a single subunit GTF and is an essential component of the PIC. It exhibits remarkable structural similarity, both at the primary and higher order structural level, among eukaryotes (Deng and Roberts 2007; Adachi et al. 2016). It is involved in initiation of transcription as well as transcription start site selection (Pinto et al. 1994; Luse 2014). It binds to the promoter region following recruitment of TFIID or SAGA complex. TFIIB interacts with both DNA elements and other proteins to facilitate transcription. It binds directly to sequences in the promoter region (Lagrange et al. 1998; Deng and Roberts 2005). The carboxy-terminal core domain of TFIIB interacts with the promoter DNA as well as TATA binding protein (TBP) and TFIIF, while the amino-terminal zinc ribbon domain interacts with the catalytic center of RNAPII (Nikolov and Burley 1997; Bushnell et al. 2004; Weinzierl and Wiesler et al. 2011; Sainsbury et al. 2013). TFIIB acts as a bridge between TFIID bound to the DNA and RNAPII (Nikolov et al. 1995). The B-finger region of TFIIB is thought to interact with the active center of the polymerase where it possibly plays a role in abortive transcription and promoter escape (Bushnell et al. 2004). The amino and carboxy-terminal domains also interact with each other to form an open or closed conformation of the protein. Thus, TFIIB can occur in an active, open conformation or a closed, inactive conformation (Glossop et al. 2004). Interaction with transcriptional activators is thought to stabilize the open form of TFIIB and thus stimulate transcription initiation (Hawkes et al. 2000; Glossop et al. 2004).
TFIIB facilitates crosstalk between 5’ and 3’ ends of a gene
Genomewide analyses have revealed that TFIIB occupies the promoter elements at the 5’ end of genes (Venters and Pugh 2013; Pugh and Venters 2016; Rossi et al. 2021). This is in accordance with the role of TFIIB in initiation of transcription. It was, however, surprising to find that TFIIB occupies the terminator region toward the 3’ end of a number of genes in budding yeast (Singh and Hampsey 2007; Mavrich et al. 2008; El Kaderi et al. 2009; Mayer et al. 2010; Medler et al. 2011; Murray et al. 2012; Al Husini et al. 2013). A similar 3’ end localization of TFIIB has been observed for a subset of genes in metazoans (Yochum et al. 2007; Mapendano et al. 2010; Henriques et al. 2012). Nearly 65% of genes exhibit crosslinking of TFIIB to both the 5’ and the 3’ end in humans (Yochum et al. 2007). The same study found TFIIB crosslinking internally to the coding region, between the 5’ and 3’ ends of genes, as well. The promoter occupancy of TFIIB is due to its interaction with the DNA elements called BREs (TFIIB Recognition Elements), which are present in the promoter region of some but not all RNAPII-transcribed genes (Deng and Roberts 2005). There is, however, no evidence for the presence of any such TFIIB-binding DNA element at the 3’ end of genes. In all likelihood, terminator localization of TFIIB may not be due to protein-DNA interaction, but rather due to protein–protein interactions of TFIIB with components of the 3’ end processing-termination machinery. This could be the reason for the failure to detect TFIIB signal at the 3’ end of genes in budding yeast by ChIP-exo approach, which is more efficient in detecting direct protein-DNA interactions (Rhee and Pugh 2012). Several independent studies have confirmed multiple interactions of TFIIB with the factors operating at the 3′ end of genes. Foremost among these interactions is with Ssu72, which is a subunit of the CPF termination complex. TFIIB exhibits both a genetic and physical interaction with Ssu72 (Pinto et al. 1994; Sun and Hampsey 1996; Wu et al. 1999; Allepuz-Fuster et al. 2019). TFIIB also interacts with the yeast CF1 subunit Rna15 as well as its human homolog CstF64 (El Kaderi et al. 2009; Wang et al. 2010). Further investigation of TFIIB-Rna15 interaction culminated in purification of a megacomplex of TFIIB with Rna14, Rna15, Pcf11, Clp1, Hrp1 and poly(A) polymerase from yeast cells (Medler et al. 2011). An independent study confirmed the existence of the interaction of TFIIB with CF1 and CPF 3’ end processing-termination complexes (Chereji et al. 2017). Further purification and characterization of the holo-TFIIB complex may reveal novel factors whose role in the transcription cycle has not yet been discovered. Taken together, these studies lead to the speculation that TFIIB functions at more than just the initiation step of transcription.
The role of TFIIB in termination of transcription
The intriguing presence of TFIIB at the 3′ end of a gene and its interaction with a gamut of termination factors raised the possibility of its involvement in termination of transcription. Indeed, TFIIB has been implicated in termination of transcription in multiple model organisms. In mammalian cells, phosphorylation of TFIIB occurs at serine-65 and is necessary for its termination function (Wang et al. 2010). TFIIB serine-65 phosphorylation regulates the TFIIB interaction with the Cstf-64 subunit of the CstF 3’ end processing-termination complex and facilitates recruitment of the termination complex near the 3’ end of a gene (Wang et al. 2010). A similar role of TFIIB in termination was demonstrated in flies and budding yeast (Henriques et al. 2012; Allepuz-Fuster et al. 2019). The sua7-1 mutant of TFIIB, in which glutamic acid is replaced by lysine at position 62, exhibits a termination defect (Allepuz-Fuster et al. 2019). In the sua7-1 mutant, promoter occupancy of TFIIB remains unaffected, but its terminator recruitment is compromised (Singh and Hampsey 2007). Furthermore, the interaction of TFIIB with the termination factors is disrupted in the sua7-1 mutant (Medler et al. 2011). Consequently, recruitment of termination factors at the 3’ end of genes is adversely affected in the sua7-1 strain leading to a termination defect (Allepuz-Fuster et al. 2019). In budding yeast, there is an alternative pathway for termination of transcription, the poly(A)-independent pathway, which requires the Nrd1-Nab1-Sen1 termination complex. TFIIB, along with the transcription activator Rap1, has been implicated in this poly(A)-independent termination as well (Roy and Chanfreau 2018).
The next obvious question is how does TFIIB facilitate termination? The simplest explanation is that TFIIB interacts with termination factors and facilitates their recruitment at the 3’ end of a gene leading to termination of transcription as described above. There is, however, an alternative or additional possibility. It is established that following initiation of transcription, TFIIB dissociates from the initiation complex. Since the B-finger/reader domain of TFIIB blocks the RNA exit channel of polymerase in the initiation complex, the release of TFIIB is critical for the polymerase to continue transcription (Kostrewa et al. 2009). A similar mechanism may explain the TFIIB function in termination as well. At the terminator, the TFIIB B-finger/reader domain may insert into the RNA exit channel of elongating RNAPII. This may block elongation thereby facilitating termination. Incidentally, glutamic acid to lysine substitution at position 62 in the sua7-1 mutant and serine-65 phosphorylation site of mammalian TFIIB, both of which affect termination, also localize to the B-reader domain (Pinto et al. 1994; Wang et al. 2010). Whether TFIIB makes physical contact with the polymerase through its B-reader domain at the 3' end, however, needs further scrutiny.
TFIIB facilitates gene looping
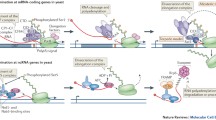
A critical question is how TFIIB, which is expected to occupy the 5′ end of a gene, is able to contact both ends of a gene. Analysis of yeast genes using the ‘Chromosome Conformation Capture’ (3C) approach revealed genes assuming a looped conformation during transcription (Ansari and Hampsey, 2005). Such gene loops are formed due to interaction of the promoter with the terminator region of a gene (Fig. 2). Our laboratory has demonstrated crucial roles of TFIIB, TFIIH, Mediator, CF1 and CPF complexes in gene looping in budding yeast (Ansari and Hampsey 2005; Medler et al. 2011; Tan-Wong et al. 2012; Al Husini et al. 2013; Mukundan and Ansari 2013; Medler and Ansari 2015). TFIIB, however, has emerged as a central player in gene loop formation (Singh and Hampsey 2007; Wang and Roberts 2010; Medler et al. 2011; Allepuz-Fuster et al. 2019). Independent studies have revealed existence of a complex of TFIIB with termination factors (Medler et al. 2011; Chereji et al. 2017). This explains how TFIIB is able to crosslink to both the 5’ and 3’ ends of a gene. Furthermore, these observations suggest that the interaction of promoter-bound TFIIB with the termination factors brings the distal ends of a gene in proximity, thereby facilitating gene loop formation.
Gene looping is completely abolished in the sua7-1 mutant of TFIIB in budding yeast (Singh and Hampsey 2007; Allepuz-Fuster 2019). In this mutant, the recruitment of TFIIB at the promoter region remains unaffected, but the 3′ end localization of the protein is almost completely abolished (Singh and Hampsey 2007). Since the TFIIB-termination factor complex is not observed in the sua7-1 mutant, we postulated that the looped gene architecture is formed primarily due to the interaction of the promoter-bound TFIIB with the termination factors occupying the 3′ end of a gene (Medler et al. 2011) (Fig. 2). In such a scenario, it is likely that the same molecule of TFIIB is crosslinked to both the ends of the gene. Consequently, TFIIB-interacting termination factors are observed at the 5′ end of a gene in wild type cells, but not in the looping-defective sua7-1 cells (Singh and Hampsey 2007).
Potential role of TFIIB in promoter directionality
Genomewide analysis of human and yeast transcription revealed a somewhat unanticipated finding that the promoter-bound RNAPII transcribes both in the sense as well as anti-sense direction (Fig. 1) (Core et al. 2008; Neil et al. 2009; Xu et al. 2009; Andersson et al. 2015; Duttke et al. 2015). Such divergent transcription from promoters has also been observed in Caenorhabditis elegans and Drosophila melanogaster (Ibrahim et al. 2018). Transcription in the downstream sense direction, which produces mRNA, continues until the polymerase reaches the 3′ end of the gene. The promoter-initiated upstream anti-sense transcription, which produces non-coding RNA, however, is terminated when the transcript is only a few hundred nucleotides long, thereby conferring directionality to the bidirectional RNAPII promoter (Fig. 1). In mammalian cells where transcription initiating in opposite directions is widespread (Vihervaara et al. 2018), asymmetric distribution of poly(A) sites and U1 snRNA-binding-sites in the promoter-proximal regions confer directionality to the promoter-bound polymerase (Almada et al. 2013; Ntini et al. 2013). In budding yeast, there is no such asymmetric distribution of poly(A) sites and U1-sites in the region surrounding the promoter. It is tempting to propose that the recruitment of termination factors in the promoter upstream region to stop uaRNA transcription is primarily dependent on gene looping in budding yeast (Tan-Wong et al. 2012; Al Husini et al. 2013). In mammalian cells, mutation of the terminator-proximal poly(A) site of the human β-globin gene results in loss of gene looping as well as promoter directionality, thereby suggesting that the looped gene architecture may also play a role in regulating promoter directionality in higher eukaryotes as well (Tan-Wong et al. 2012). Since TFIIB is a central player in gene loop formation, we propose that, at least in budding yeast, TFIIB indirectly influences promoter directionality by affecting gene loop formation (Tan-Wong et al. 2012; Al Husini et al. 2013; Agarwal and Ansari 2016). Consequently, the looping-defective sua7-1 mutant of TFIIB exhibits loss of promoter directionality (Tan-Wong et al. 2012; Agarwal and Ansari 2016). TFIIB is not the only transcription factor to play a role in promoter directionality, as the transcription factor Rap1 has also been implicated in repressing divergent transcription in yeast (Wu and Van Werven 2019; Rossi et al. 2021). A recent study in plants also implicated gene looping in promoter directionality similar to that in yeast (Gagliardi et al. 2019). Whether plant gene looping is dependent on TFIIB, however, needs further scrutiny.
TFIIB involvement in viral pathogenesis
The multifaceted role of TFIIB in eukaryotic transcription extends to its emergence as a critical player in viral pathogenesis. TFIIB has been found crucial to pathogenesis of many human and animal viruses including human immunodeficiency virus, herpes simplex virus, vaccinia virus, Thogoto virus, hepatitis virus, Epstein-Barr virus and gammaherpesviruses (O’Brien and Ansari 2021). A recent computational analyses revealed TFIIB to be a high confidence transcriptional target of coronavirus as well (Ochsner et al. 2020). Viruses have to transcribe their genes to produce proteins to complete their life cycle and infect new hosts (An et al. 2019; Liu et al. 2020). To accomplish this, viruses utilize host transcription machinery, and specifically have been shown to hijack host TFIIB (Agostini et al. 1996; Gelev et al. 2014; Liu et al. 2020). Key to the viral targeting of TFIIB are viral transcriptional regulatory proteins, which directly interact with host TFIIB to accomplish expression of their genes and to repress transcription of host antiviral genes, especially those linked to the antiviral immune response (Gelev et al. 2014; Haas et al. 2018; Yang and You, 2020; Wang et al. 2020). Some viruses have evolved transcription regulatory proteins with a three-dimensional structure very similar to TFIIB, demonstrating the importance of TFIIB for viral persistence (Grimm et al. 2019; Hillen et al. 2019; Liu et al. 2020; Cackett et al. 2020). TFIIB-like viral proteins of vaccinia virus and African Swine Fever Virus exhibit similarities with host TFIIB in crucial and conserved regions such as the B-ribbon, B-cyclin, and B-reader domains (Grimm et al. 2019; Cackett et al. 2020). These conserved regions of TFIIB may be crucial for facilitating viral transcription. Whether targeting of TFIIB by viral transcriptional regulators affects the initiation step only, or termination and promoter directionality are also impacted, needs further investigation. It is highly likely that the monomeric nature, involvement at early steps in PIC assembly and pleiotropic role of TFIIB in transcription cycle makes it a preferred target of viruses during pathogenesis. The nature of viral utilization of TFIIB for expression of its own genes, along with selective repression of host antiviral genes, makes TFIIB a potential candidate for antiviral therapies.
Future perspectives
Since its discovery as a GTF, TFIIB has primarily been thought of as a factor limited to involvement in PIC formation and initiation of transcription. Recent studies presented here have challenged this dogma. The role of TFIIB in the transcription cycle extends beyond the initiation step. Different steps of transcription do not occur separately as discreet events, but rather intertwine with each other. It is therefore not surprising that TFIIB, in addition to its established role in initiation, also impacts termination, gene loop formation and promoter directionality (Fig. 1). Further studying TFIIB in the context of these newly discovered roles will augment our understanding of the process of transcription.
The recently observed roles of TFIIB in termination and promoter directionality are dependent to a great extent on its ability to simultaneously contact the distal ends of a gene and transform a transcribing linear gene into a looped structure. At the core of the gene loop is the complex of TFIIB with termination factors (Fig. 2). This complex called the holo-TFIIB complex is transiently formed during transcription and can be purified only from transcriptionally active chromatin (Chereji et al. 2017). The characterization of this complex may reveal novel accessory factors whose role in the transcription cycle, especially in termination, reinitiation and promoter directionality has so far eluded us. It will be worth investigating if such a complex is unique to yeast or if it is a general feature of eukaryotic transcription machinery.
The process of transcription is essential to human life but is also necessary for viruses to replicate and infect a new host. TFIIB has been found crucial for pathogenesis of many deadly viruses, as viral transcription is often dependent upon host TFIIB. The importance of TFIIB as a potential antiviral target has been made apparent as, in some cases, inactivation of host TFIIB severely impairs viral replication without adversely affecting general host transcription (reviewed in O’Brien and Ansari 2021). A thorough examination of the role of TFIIB in the transcription cycle will not only help understand the process of transcription in its entirety but may also help to find targets for antiviral therapies.
References
Adachi N, Senda T, Horikoshi M (2016) Uncovering ancient transcription systems with a novel evolutionary indicator. Sci Rep 6(1):1–12
Agarwal N, Ansari A (2016) Enhancement of transcription by a splicing-competent intron is dependent on promoter directionality. PLoS Genet 12(5):e1006047
Agostini I, Navarro JM, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J (1996) The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J Mol Biol 261:599–606
Al Husini N, Kudla P, Ansari A (2013) A role for CF1A 3′ end processing complex in promoter-associated transcription. PLoS Genet 9(8):e1003722
Al-Husini N, Medler S, Ansari A (2020) Crosstalk of promoter and terminator during RNA polymerase II transcription cycle. Biochim Biophys Acta Gene Regul Mech 2:194657
Allepuz-Fuster P, O’Brien MJ, González-Polo N, Pereira B, Dhoondia Z, Ansari A, Calvo O (2019) RNA polymerase II plays an active role in the formation of gene loops through the Rpb4 subunit. Nucleic Acids Res 47(17):8975–8987. https://doi.org/10.1093/nar/gkz597
Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA (2013) Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499(7458):360–363
An P, Guo JT, Winkler CA (2019) Host genetics in viral pathogenesis and control. Front Genet 10:1038
Andersson R, Sandelin A, Danko CG (2015) A unified architecture of transcriptional regulatory elements. Trends Genet 31(8):426–433
Ansari A, Hampsey M (2005) A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19(24):2969–2978
Baejen C, Andreani J, Torkler P, Battaglia S, Schwalb B, Lidschreiber M, Cramer P (2017) Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol Cell 66(1):38–49
Bushnell DA, Westover KD, Davis RE, Kornberg RD (2004) Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 45 Angstroms. Science 303(5660):983–988
Cackett G, Matelska D, Sýkora M, Portugal R, Malecki M, Bähler J, Werner F (2020) The African swine fever virus transcriptome. J Virol 94(9):e00119-e120
Chereji RV, Bharatula V, Elfving N, Blomberg J, Larsson M, Morozov AV, Björklund S (2017) Mediator binds to boundaries of chromosomal interaction domains and to proteins involved in DNA looping, RNA metabolism, chromatin remodeling, and actin assembly. Nucleic Acids Res 45(15):8806–8821
Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848
Deng W, Roberts SG (2005) A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev 19(20):2418–2423
Deng W, Roberts SG (2007) TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma 116(5):417–429
Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Ohler U (2015) Perspectives on unidirectional versus divergent transcription. Mol Cell 60(3):348–349
El Kaderi B, Medler S, Raghunayakula S, Ansari A (2009) Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem 284(37):25015–25025. https://doi.org/10.1074/jbc.M109.007948
Gagliardi D, Cambiagno DA, Arce AL, Tomassi AH, Giacomelli JI, Ariel FD, Manavella PA (2019) Dynamic regulation of chromatin topology and transcription by inverted repeat-derived small RNAs in sunflower. Proc Natl Acad Sci 116(35):17578–17583
Gelev V, Zabolotny JM, Lange M, Hiromura M, Yoo SW, Orlando JS, Usheva A (2014) A new paradigm for transcription factor TFIIB functionality. Sci Rep 4(1):1–8
Glossop JA, Dafforn TR, Roberts SGE (2004) A conformational change in TFIIB is required for activor-mediated assembly of the preinitiation complex. Nucleic Acids Res 32:1829–1835
Grimm C, Hillen HS, Bedenk K, Bartuli J, Neyer S, Zhang Q, Fischer U (2019) Structural basis of poxvirus transcription: vaccinia RNA polymerase complexes. Cell 179(7):1537–1550
Haas DA, Meiler A, Geiger K, Vogt C, Preuss E, Kochs G, Pichlmair A (2018) Viral targeting of TFIIB impairs de novo polymerase II recruitment and affects antiviral immunity. PLoS Pathog 14(4):e1006980
Hahn S, Young ET (2011) Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189(3):705–736
Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN (2002) Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol 22(18):6441–6457
Hawkes NA, Evans R, Roberts SG (2000) The conformation of the transcription factor TFIIB modulates the response to transcriptional activators in vivo. Curr Biol 10(5):273–276
Henriques T, Ji Z, Tan-Wong SM, Carmo AM, Tian B, Proudfoot NJ et al (2012) Transcription termination between polo and snap, two closely spaced tandem genes of D. melanogaster. Transcription. 3(4):198–212
Hillen HS, Bartuli J, Grimm C, Dienemann C, Bedenk K, Szalay AA, Cramer P (2019) Structural basis of poxvirus transcription: transcribing and capping vaccinia complexes. Cell 179(7):1525–1536
Ibrahim MM, Karabacak A, Glahs A, Kolundzic E, Hirsekorn A, Carda A, Ohler U (2018) Determinants of promoter and enhancer transcription directionality in metazoans. Nat Commun 9(1):1–15
Jonkers I, Lis JT (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16(3):167–177
Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P (2009) RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462(7271):323–330
Krishnamurthy S, Hampsey M (2009) Eukaryotic transcription initiation. Curr Biol 19(4):R153–R156
Kuehner JN, Pearson EL, Moore C (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12(5):283–294
Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH (1998) New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev 12(1):34–44
Liu X, Hong T, Parameswaran S, Ernst K, Marazzi I, Weirauch MT, Bass JIF (2020) Human virus transcriptional regulators. Cell 182(1):24–37
Luse DS (2014) The RNA polymerase II preinitiation complex: through what pathway is the complex assembled? Transcription 5(1):e27050
Lykke-Andersen S, Mapendano CK, Jensen TH (2011) An ending is a new beginning: transcription termination supports re-initiation. Cell Cycle 10:863–865
Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH (2010) Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40(3):410–422
Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Franklin Pugh B (2008) A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18:1073–1083
Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17(10):1272–1278
Medler S, Ansari A (2015) Gene looping facilitates TFIIH kinase-mediated termination of transcription. Sci Rep 5(1):1–11
Medler S, Al-Husini N, Raghunayakula S, Mukundan B, Aldea A, Ansari A (2011) Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J Biol Chem 286(39):33709–33718
Mischo HE, Proudfoot NJ (2013) Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta Gene Regul Mech 1829(1):174–185
Mukundan B, Ansari A (2013) Srb5/Med18-mediated termination of transcription is dependent on gene looping. J Biol Chem 288(16):11384–11394
Murray SC, Serra Barros A, Brown DA, Dudek P, Ayling J, Mellor J (2012) A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res 40(6):2432–2444
Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457(7232):1038–1042
Nikolov DB, Burley SK (1997) RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci 94(1):15–22
Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Burley SK (1995) Crystal structure of a TFIIB–TBP–TATA-element ternary complex. Nature 377(6545):119–128
Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, Jensen TH (2013) Polyadenylation site–induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol 20(8):923–928
O’Brien MJ, Ansari A (2021) Critical involvement of TFIIB in viral pathogenesis. Front Mol Biosci 8:308
Ochsner SA, Pillich RT, McKenna NJ (2020) Consensus transcriptional regulatory networks of coronavirus-infected human cells. Sci Data 7(1):1–20
Pinto I, Wu WH, Na JG, Hampsey M (1994) Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem 269(48):30569–30573
Pugh BF, Venters BJ (2016) Genomic organization of human transcription initiation complexes. PLoS ONE 11(2):e0149339
Rhee HS, Pugh BF (2012) Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483(7389):295–301
Richard P, Manley JL (2009) Transcription termination by nuclear RNA polymerases. Genes Dev 23(11):1247–1269
Rossi MJ, Kuntala PK, Lai WK, Yamada N, Badjatia N, Mittal C, Pugh BF (2021) A high-resolution protein architecture of the budding yeast genome. Nature 592(7853):309–314
Roy K, Chanfreau GF (2018) A global function for transcription factors in assisting RNA polymerase II termination. Transcription 9(1):41–46
Sainsbury S, Niesser J, Cramer P (2013) Structure and function of the initially transcribing RNA polymerase II–TFIIB complex. Nature 493(7432):437–440
Singh BN, Hampsey M (2007) A transcription-independent role for TFIIB in gene looping. Mol Cell 27:806–816
Sun ZW, Hampsey M (1996) Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol 16(4):1557–1566
Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE et al (2012) Gene loops enhance transcriptional directionality. Science 338(6107):671–675
Venters BJ, Pugh BF (2013) Genomic organization of human transcription initiation complexes. Nature 502(7469):53–58
Vihervaara A, Duarte FM, Lis JT (2018) Molecular mechanisms driving transcriptional stress responses. Nat Rev Genet 19(6):385–397
Wang Y, Roberts SG (2010) New insights into the role of TFIIB in transcription initiation. Transcription 1(3):126–129
Wang Y, Fairley JA, Roberts SG (2010) Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol 20(6):548–553
Wang X, Hennig T, Whisnant AW, Erhard F, Prusty BK, Friedel CC, Shi Y (2020) Herpes simplex virus blocks host transcription termination via the bimodal activities of ICP27. Nat Commun 11(1):1–13
Weinzierl RO, Wiesler SC (2011) Revealing the functions of TFIIB. Transcription 2(6):254–257
Woychik NA, Hampsey M (2002) The RNA polymerase II machinery: structure illuminates function. Cell 108(4):453–463
Wu AC, Van Werven FJ (2019) Transcribe this way: Rap1 confers promoter directionality by repressing divergent transcription. Transcription 10(3):164–170
Wu WH, Pinto I, Chen BS, Hampsey M (1999) Mutational analysis of yeast TFIIB: a functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153(2):643–652
Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457(7232):1033–1037
Yang JF, You J (2020) Regulation of polyomavirus transcription by viral and cellular factors. Viruses 12(10):1072
Yochum GS, Rajarama V, Cleland R, McWeeney S (2007) Localization of TFIIB binding regions using serial analysis of chromatin occupancy. MBC Mol Biol 8:102
Yudkovsky N, Ranish JA, Hahn S (2000) A transcription reinitiation intermediate that is stabilized by activator. Nature 408(6809):225–229
Acknowledgements
We thank Dr. Lori Pile of Wayne State University for careful reading of the manuscript and giving valuable suggestions.
Funding
This research was supported by NSF Grant MCB-1936030.
Author information
Authors and Affiliations
Contributions
AA conceptualized and edited the manuscript. MJO wrote the original draft. Both authors contributed to the article and approve the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Communicated by Michael Polymenis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
O’Brien, M.J., Ansari, A. Beyond the canonical role of TFIIB in eukaryotic transcription. Curr Genet 68, 61–67 (2022). https://doi.org/10.1007/s00294-021-01223-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-021-01223-x