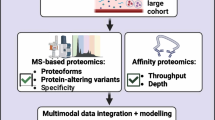
Abstract
Proteomics, the study of proteins and their functions, has greatly evolved due to advances in analytical chemistry and computational biology. Unlike genomics or transcriptomics, proteomics captures the dynamic and diverse nature of proteins, which play crucial roles in cellular processes. This is exemplified in cancer, where genomic and transcriptomic information often falls short in reflecting actual protein expression and interactions. Liquid chromatography–mass spectrometry (LC-MS) is pivotal in proteomic data generation, enabling high-throughput analysis of protein samples. The MS-based workflow involves protein digestion, chromatographic separation, ionization, and fragmentation, leading to peptide identification and quantification. Computational biostatistics, particularly using tools in R (R Foundation for Statistical Computing, Vienna, Austria; www.R-project.org), aid in data analysis, revealing protein expression patterns and correlations with clinical variables. Proteomic studies can be explorative, aiming to characterize entire proteomes, or targeted, focusing on specific proteins of interest. The integration of proteomics with genomics addresses database limitations and enhances peptide identification. Case studies in intrahepatic cholangiocarcinoma, glioblastoma multiforme, and pancreatic ductal adenocarcinoma highlight proteomics’ clinical applications, from subtyping cancers to identifying diagnostic markers. Moreover, proteomic data augment molecular tumor boards by providing deeper insights into pathway activities and genomic mutations, supporting personalized treatment decisions. Overall, proteomics contributes significantly to advancing our understanding of cellular biology and improving clinical care.
Zusammenfassung
Proteomik beschäftigt sich mit der globalen Analyse aller Proteine einer Probe und hat sich in den letzten Jahren enorm weiterentwickelt. Proteine durchlaufen vielfältige Modifikations‑, Transport- und Abbauprozesse, derentwegen das Proteom hochdynamisch ist. Ein Beispiel dafür sind Krebserkrankungen, bei denen genomische und transkriptomische Informationen die tatsächliche Proteinexpression und -interaktion kaum widerspiegeln können. Die an Flüssigchromatographie gekoppelte Massenspektrometrie (LC-MS) ist von zentraler Bedeutung bei der Generierung proteomischer Daten und ermöglicht die Hochdurchsatzanalyse zur Peptididentifizierung und -quantifizierung im Massenspektrometer aus sehr geringen Materialmengen. Computergestützte Biostatistik, insbesondere unter Verwendung von R (R Foundation for Statistical Computing, Wien, Österreich; www.R-project.org), hilft bei der Datenanalyse und deckt Proteinexpressionsmuster und Korrelationen mit klinischen Variablen auf. Proteomische Studien können explorativ sein und auf die Charakterisierung ganzer Proteome abzielen oder zielgerichtet sein und sich auf bestimmte interessierende Proteine konzentrieren. Die Integration von Proteomik in Genomik zielt auf die Beseitigung von Einschränkungen in Datenbanken ab und verbessert die Peptididentifizierung. Verschiedene Fallstudien verdeutlichen die klinischen Anwendungen der Proteomik, von der Subtypisierung von Krebserkrankungen bis zur Identifizierung diagnostischer Marker. Darüber hinaus unterstützen proteomische Daten molekulare Tumorboards, indem sie tiefere Einblicke in Signalwegaktivitäten und genomische Mutationen liefern und so bei personalisierten Behandlungsentscheidungen helfen. Insgesamt trägt die Proteomik wesentlich dazu bei, unser Verständnis der Zell- und Tumorbiologie zu erweitern und die klinische Versorgung zu verbessern.




Similar content being viewed by others
References
Aebersold R, Mann M (2016) Mass-spectrometric exploration of proteome structure and function. Nature 537:347–355
Banales JM, Marin JJG, Lamarca A et al (2020) Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol 17:557–588
Bhowmick NA, Moses HL (2005) Tumor–stroma interactions. Curr Opin Genet Dev 15:97–101
Chen C, Hou J, Tanner JJ, Cheng J (2020) Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci 21:2873
Chen G, Gharib TG, Huang C‑C et al (2002) Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 1:304–313
Coscia F, Lengyel E, Duraiswamy J et al (2018) Multi-level Proteomics identifies CT45 as a chemosensitivity mediator and immunotherapy target in ovarian cancer. Cell 175:159–170.e16
Demichev V, Szyrwiel L, Yu F et al (2022) dia-PASEF data analysis using fragpipe and DIA-NN for deep proteomics of low sample amounts. Nat Commun 13:3944
Doll S, Kriegmair MC, Santos A et al (2018) Rapid proteomic analysis for solid tumors reveals LSD1 as a drug target in an end-stage cancer patient. Mol Oncol 12:1296–1307
Doroshow DB, Bhalla S, Beasley MB et al (2021) PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol 18:345–362
Fröhlich K, Brombacher E, Fahrner M et al (2022) Benchmarking of analysis strategies for data-independent acquisition proteomics using a large-scale dataset comprising inter-patient heterogeneity. Nat Commun 13:2622
Geyer PE, Holdt LM, Teupser D, Mann M (2017) Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol 13:942
Gillet LC, Leitner A, Aebersold R (2016) Mass spectrometry applied to bottom-up proteomics: entering the high-throughput era for hypothesis testing. Annu Rev Anal Chem 9:449–472
Hoefflin R, Geißler A‑L, Fritsch R et al (2018) Personalized clinical decision making through implementation of a molecular tumor board: a German single-center experience. JCO Precis Oncol (1–16)
Jones DTW, Kocialkowski S, Liu L et al (2008) Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68:8673–8677
Landrum MJ, Lee JM, Riley GR et al (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:D980–D985
Larson KL, Huang B, Weiss HL et al (2021) Clinical outcomes of molecular tumor boards: a systematic review. JCO Precis Oncol (1122–1132)
Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550
Luchini C, Lawlor RT, Milella M, Scarpa A (2020) Molecular tumor boards in clinical practice. Trends Cancer 6:738–744
Mani DR, Krug K, Zhang B et al (2022) Cancer proteogenomics: current impact and future prospects. Nat Rev Cancer 22:298–313
Organisation mondiale de la santé (2021) Centre international de recherche sur le cancer. In: Central nervous system tumours, 5th edn. International agency for research on cancer, Lyon
Santos A, Colaço AR, Nielsen AB et al (2022) A knowledge graph to interpret clinical proteomics data. Nat Biotechnol 40:692–702
Schwaederle M, Parker BA, Schwab RB et al (2014) Molecular tumor board: the university of california san diego moores cancer center experience. The Oncol 19:631–636
Schwanhäusser B, Busse D, Li N et al (2011) Global quantification of mammalian gene expression control. Nature 473:337–342
Sharma K, D’Souza RCJ, Tyanova S et al (2014) Ultradeep human phosphoproteome reveals a distinct regulatory nature of tyr and Ser/Thr-based signaling. Cell Rep 8:1583–1594
Tan HT, Lee YH, Chung MCM (2012) Cancer proteomics: CANCER PROTEOMICS. Mass Spectrom Rev 31:583–605
Vidova V, Spacil Z (2017) A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition. Anal Chim Acta 964:7–23
Werner J, Bernhard P, Cosenza-Contreras M et al (2023) Targeted and explorative profiling of kallikrein proteases and global proteome biology of pancreatic ductal adenocarcinoma, chronic pancreatitis, and normal pancreas highlights disease-specific proteome remodelling. Neoplasia 36:100871
Witze ES, Old WM, Resing KA, Ahn NG (2007) Mapping protein post-translational modifications with mass spectrometry. Nat Methods 4:798–806
Zhu Y, Aebersold R, Mann M, Guo T (2021) SnapShot: Clinical proteomics. Cell 184:4840–4840.e1
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
T. Werner, M. Fahrner, and O. Schilling declare that they have no competing interests.
All human studies described were carried out with the approval of the responsible ethics committees, in accordance with national law and in accordance with the Declaration of Helsinki of 1975 (as amended). A declaration of consent was provided from all patients involved. All national guidelines for keeping and handling laboratory animals have been adhered to and the necessary approvals from the responsible authorities have been obtained.
The supplement containing this article is not sponsored by industry.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.

Scan QR code & read article online
Rights and permissions
About this article
Cite this article
Werner, T., Fahrner, M. & Schilling, O. Using proteomics for stratification and risk prediction in patients with solid tumors. Pathologie 44 (Suppl 3), 176–182 (2023). https://doi.org/10.1007/s00292-023-01261-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00292-023-01261-x