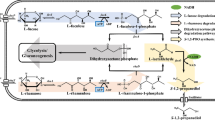
Abstract
Metabolic engineering provides a more sustainable alternative by using complex metabolic processes to generate valuable compounds by exploiting nature's metabolic networks. Metabolic flux analysis is frequently used in metabolic engineering to improve production yields and selectivity of compounds including biofuels, medicines, and industrial chemicals. It analyzes metabolic pathways and flux distribution to identify essential steps and targets for optimization, mainly to increase biopolymer synthesis, including polyhydroxybutyrate (PHB). PHB is a significant carbon reservoir due to its capacity to store carbon in a biodegradable polymer form in sectors such as packaging, biomedicine, and environmental remediation, making it an economically and environmentally friendly substitute to existing nonrenewable resources. Metabolic flux analysis and metabolic engineering are essential in synthesizing PHB, a biodegradable polymer with several uses. By combining these techniques, researchers can enhance efficiency, boost yields, and develop microorganisms with improved PHB synthesis capabilities. Metabolic flux analysis can identify rate-limiting steps in the PHB biosynthesis pathway and evaluate flux distribution, enabling the identification of metabolic engineering targets that may increase PHB production. Metabolic engineering may also increase the expression of PHB biosynthesis genes or add new genes to the pathway. The study analyzed here examines the significance of combining metabolic flux analysis and metabolic engineering, as well as their potential to considerably increase PHB production and improve the economic feasibility of this key biopolymer.

Similar content being viewed by others
References
Libourel IGL, Shachar-Hill Y (2008) Metabolic flux analysis in plants: from intelligent design to rational engineering. Annu Rev Plant Biol 59:625–650. https://doi.org/10.1146/annurev.arplant.58.032806.103822
Boghigian BA, Seth G, Kiss R, Pfeifer BA (2010) Metabolic flux analysis and pharmaceutical production. Metab Eng 12:81–95. https://doi.org/10.1016/j.ymben.2009.10.004
Antoniewicz MR (2013) 13C metabolic flux analysis: Optimal design of isotopic labeling experiments. Curr Opin Biotechnol 24:1116–1121. https://doi.org/10.1016/j.copbio.2013.02.003
Schwender J, Ohlrogge J, Shachar-Hill Y (2004) Understanding flux in plant metabolic networks. Curr Opin Plant Biol 7:309–317. https://doi.org/10.1016/j.pbi.2004.03.016
Copeland WB, Bartley BA, Chandran D et al (2012) Computational tools for metabolic engineering. Metab Eng 14:270–280. https://doi.org/10.1016/j.ymben.2012.03.001
Lee SY, Park JM, Kim TY (2011) Application of metabolic flux analysis in metabolic engineering. Methods Enzymol 498:67–93. https://doi.org/10.1016/B978-0-12-385120-8.00004-8
Antoniewicz MR (2013) Dynamic metabolic flux analysis-tools for probing transient states of metabolic networks. Curr Opin Biotechnol 24:973–978. https://doi.org/10.1016/j.copbio.2013.03.018
Kumar RR, Prasad S (2011) Metabolic engineering of bacteria. Indian J Microbiol 51:403–409. https://doi.org/10.1007/s12088-011-0172-8
Chen X, Zhou L, Tian K et al (2013) Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv 31:1200–1223. https://doi.org/10.1016/j.biotechadv.2013.02.009
Gao J, Du M, Zhao J et al (2022) Design of a genetically encoded biosensor to establish a high-throughput screening platform for L-cysteine overproduction. Metab Eng 73:144–157. https://doi.org/10.1016/j.ymben.2022.07.007
Liu R, Bassalo MC, Zeitoun RI, Gill RT (2015) Genome scale engineering techniques for metabolic engineering. Metab Eng 32:143–154. https://doi.org/10.1016/j.ymben.2015.09.013
Wang Y, Wondisford FE, Song C et al (2020) Metabolic flux analysis—linking isotope labeling and metabolic fluxes. Metabolites 10:1–21. https://doi.org/10.3390/metabo10110447
Metallo CM, Walther JL, Stephanopoulos G (2009) Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol 144:167–174. https://doi.org/10.1016/j.jbiotec.2009.07.010
Orman MA, Berthiaume F, Androulakis IP, Ierapetritou MG (2011) Advanced stoichiometric analysis of metabolic networks of mammalian systems. Crit Rev Biomed Eng 39:511–534. https://doi.org/10.1615/CritRevBiomedEng.v39.i6.30
Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotechnol 28:245–248. https://doi.org/10.1038/nbt.1614
Sun S, Ding Y, Liu M et al (2020) Comparison of glucose, acetate and ethanol as carbon resource for production of poly(3-Hydroxybutyrate) and other Acetyl-CoA derivatives. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2020.00833
Wang Q, Liu C, Xian M et al (2012) Biosynthetic pathway for poly(3-Hydroxypropionate) in recombinant Escherichia coli. J Microbiol 50:693–697. https://doi.org/10.1007/s12275-012-2234-y
Liu XW, Wang HH, Chen JY et al (2009) Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by recombinant Escherichia coli harboring propionyl-CoA synthase gene (prpE) or propionate permease gene (prpP). Biochem Eng J 43:72–77. https://doi.org/10.1016/j.bej.2008.09.001
Taguchi K, Aoyagi Y, Matsusaki H et al (1999) Over-expression of 3-ketoacyl-ACP synthase III or malonyl-CoA-ACP transacylase gene induces monomer supply for polyhydroxybutyrate production in Escherichia coli HB101. Biotechnol Lett 21:579–584. https://doi.org/10.1023/A:1005572526080
Kim M, Baek J, Lee J (2006) Comparison of H2H2 accumulation by Rhodobacter sphaeroides KD131 and its uptake hydrogenase and PHB synthase deficient mutant. Int J Hydrog Energy 31:121–127. https://doi.org/10.1016/j.ijhydene.2004.10.023
Wang Z, Qin W, Bao S et al (2013) The influences of aerobic and anaerobic conditions on PHB and glycerin yields in the process of lignin degradation by Pseudomonas stutzeri P156. Adv Mater Res 634–638:1170–1174. https://doi.org/10.4028/www.scientific.net/AMR.634-638.1170
Towijit U, Songruk N, Lindblad P et al (2018) Co-overexpression of native phospholipid-biosynthetic genes plsX and plsC enhances lipid production in Synechocystis sp. PCC 6803. Sci Rep. https://doi.org/10.1038/s41598-018-31789-5
Muller EEL, Sheik AR, Wilmes P (2014) Lipid-based biofuel production from wastewater. Curr Opin Biotechnol 30:9–16. https://doi.org/10.1016/j.copbio.2014.03.007
Haywood GW, Anderson AJ, Chu L, Dawes EA (1988) The role of NADH- and NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol Lett 52:259–264. https://doi.org/10.1111/j.1574-6968.1988.tb02607.x
Gradinaru VR (2005) Acyl-CoA dehydrogenases: mechanistic studies on medium chain Acyl-CoA dehydrogenase dissertation. Bach
Tan HT, Chek MF, Miyahara Y et al (2022) Characterization of an (R)-specific enoyl-CoA hydratase from streptomyces sp. strain CFMR 7: A metabolic tool for enhancing the production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate). J Biosci Bioeng 134:288–294. https://doi.org/10.1016/j.jbiosc.2022.07.005
Volodina E, Steinbüchel A (2014) (S)-3-hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase (FadB’) from fatty acid degradation operon of Ralstonia eutropha H16. AMB Express 4:1–9. https://doi.org/10.1186/s13568-014-0069-0
Segura D, Vargas E, Espín G (2000) β-Ketothiolase genes in Azotobacter vinelandii. Gene 260:113–120. https://doi.org/10.1016/S0378-1119(00)00462-5
Tang R, Peng X, Weng C, Han Y (2022) The overexpression of phasin and regulator genes promoting the synthesis of polyhydroxybutyrate in cupriavidus necator H16 under nonstress conditions. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01458-21
Tang R, Weng C, Peng X, Han Y (2020) Metabolic engineering of Cupriavidus necator H16 for improved chemoautotrophic growth and PHB production under oxygen-limiting conditions. Metab Eng 61:11–23. https://doi.org/10.1016/j.ymben.2020.04.009
Aristya GR, Lin YJ, Chang JS et al (2022) Polyhydroxybutyrate (PHB) production from crude glycerol by genetic engineering of Rhodotorula glutinis. Bioresour Technol Rep. https://doi.org/10.1016/j.biteb.2022.101048
John ME, Keller G (1996) Metabolic pathway engineering in cotton: Biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci U S A 93:12768–12773. https://doi.org/10.1073/pnas.93.23.12768
Koch M, Bruckmoser J, Scholl J et al (2020) Maximizing PHB content in Synechocystis sp. PCC 6803: a new metabolic engineering strategy based on the regulator PirC. Microb Cell Fact. https://doi.org/10.1186/s12934-020-01491-1
Matsumoto K, Morimoto K, Gohda A et al (2011) Improved polyhydroxybutyrate (PHB) production in transgenic tobacco by enhancing translation efficiency of bacterial PHB biosynthetic genes. J Biosci Bioeng 111:485–488. https://doi.org/10.1016/j.jbiosc.2010.11.020
Müller-Santos M, Koskimäki JJ, Alves LPS et al (2021) The protective role of PHB and its degradation products against stress situations in bacteria. FEMS Microbiol Rev. https://doi.org/10.1093/femsre/fuaa058
Nishihata S, Kondo T, Tanaka K et al (2018) Bradyrhizobium diazoefficiens USDA110 PhaR functions for pleiotropic regulation of cellular processes besides PHB accumulation. BMC Microbiol. https://doi.org/10.1186/s12866-018-1317-2
Lee TR, Lin JS, Wang SS, Shaw GC (2004) PhaQ, a new class of poly-β-hydroxybutyrate (PHB)-responsive repressor, regulates phaQ and phaP (Phasin) expression in Bacillus megaterium through Interaction with PHB. J Bacteriol 186:3015–3021. https://doi.org/10.1128/JB.186.10.3015-3021.2004
Pfeiffer D, Jendrossek D (2014) PhaM Is the Physiological Activator of Poly(3-Hydroxybutyrate) (PHB) Synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol 80:555–563. https://doi.org/10.1128/AEM.02935-13
Lee YR, Nur Fitriana H, Lee SY et al (2020) Molecular profiling and optimization studies for growth and phb production conditions in Rhodobacter sphaeroides. Energies. https://doi.org/10.3390/en13236471
Xie J, Zhou J, Zhang H, Li Y (2011) Increasing reductant NADPH content via metabolic engineering of PHB synthesis pathway in Synechocystis sp. PCC 6803. Shengwu Gongcheng Xuebao/Chin J Biotechnol 27:998–1004
Park YL, Song HS, Choi TR et al (2021) Revealing of sugar utilization systems in Halomonas sp. YLGW01 and application for poly(3-hydroxybutyrate) production with low-cost medium and easy recovery. Int J Biol Macromol 167:151–159. https://doi.org/10.1016/j.ijbiomac.2020.11.163
Tumlirsch T, Sznajder A, Jendrossek D (2015) Formation of polyphosphate by polyphosphate kinases and its relationship to poly(3-hydroxybutyrate) accumulation in Ralstonia eutropha strain H16. Appl Environ Microbiol 81:8277–8293. https://doi.org/10.1128/AEM.02279-15
Handrick R, Reinhardt S, Kimmig P, Jendrossek D (2004) The “intracellular” poly(3-hydroxybutyrate) (PHB) depolymerase of Rhodospirillum rubrum is a periplasm-located protein with specificity for native PHB and with structural similarity to extracellular PHB depolymerases. J Bacteriol 186:7243–7253. https://doi.org/10.1128/JB.186.21.7243-7253.2004
Kobayashi T, Shiraki M, Abe T et al (2003) Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J Bacteriol 185:3485–3490. https://doi.org/10.1128/JB.185.12.3485-3490.2003
de las Heras AM, Portugal-Nunes DJ, Rizza N et al (2016) Anaerobic poly-3-d-hydroxybutyrate production from xylose in recombinant Saccharomyces cerevisiae using a NADH-dependent acetoacetyl-CoA reductase. Microb Cell Fact. https://doi.org/10.1186/s12934-016-0598-0
Shi LL, Da YY, Zheng WT et al (2020) Production of polyhydroxyalkanoate from acetate by metabolically engineered Aeromonas hydrophilia. J Biosci Bioeng 130:290–294. https://doi.org/10.1016/j.jbiosc.2020.05.003
Poole P, Allaway D (2000) Carbon and nitrogen metabolism in Rhizobium. In: Advances in microbial physiology. pp 117–163
Lin Z, Zhang Y, Yuan Q et al (2015) Metabolic engineering of Escherichia coli for poly(3-hydroxybutyrate) production via threonine bypass. Microb Cell Fact 14:185. https://doi.org/10.1186/s12934-015-0369-3
Park S, Kim GB, Kim HU et al (2019) Enhanced production of poly-3-hydroxybutyrate (PHB) by expression of response regulator DR1558 in recombinant Escherichia coli. Int J Biol Macromol 131:29–35. https://doi.org/10.1016/j.ijbiomac.2019.03.044
Ku JT, Chen AY, Lan EI (2020) Metabolic engineering design strategies for increasing acetyl-coa flux. Metabolites. https://doi.org/10.3390/metabo10040166
Yishai O, Goldbach L, Tenenboim H et al (2017) Engineered assimilation of exogenous and endogenous formate in Escherichia coli. ACS Synth Biol 6:1722–1731. https://doi.org/10.1021/acssynbio.7b00086
Zhao Y, Jian X, Wu J et al (2019) Elucidation of the biosynthesis pathway and heterologous construction of a sustainable route for producing umbelliferone. J Biol Eng. https://doi.org/10.1186/s13036-019-0174-3
Kocharin K, Chen Y, Siewers V, Nielsen J (2012) Engineering of acetyl-CoA metabolism for the improved production of polyhydroxybutyrate in Saccharomyces cerevisiae. AMB Express. https://doi.org/10.1186/2191-0855-2-52
Edwards CB, Copes N, Brito AG et al (2013) Malate and Fumarate Extend Lifespan in Caenorhabditis elegans. PLoS ONE. https://doi.org/10.1371/journal.pone.0058345
Kabir MM, Shimizu K (2003) Gene expression patterns for metabolic pathway in pgi knockout Escherichia coli with and without phb genes based on RT-PCR. J Biotechnol 105:11–31. https://doi.org/10.1016/S0168-1656(03)00170-6
Naranjo JM, Posada JA, Higuita JC, Cardona CA (2013) Valorization of glycerol through the production of biopolymers: The PHB case using Bacillus megaterium. Bioresour Technol 133:38–44. https://doi.org/10.1016/j.biortech.2013.01.129
de Souza L, Manasa Y, Shivakumar S (2020) Bioconversion of lignocellulosic substrates for the production of polyhydroxyalkanoates. Biocatal Agric Biotechnol. https://doi.org/10.1016/j.bcab.2020.101754
Johnson K, Kleerebezem R, van Loosdrecht MCM (2010) Influence of the C/N ratio on the performance of polyhydroxybutyrate (PHB) producing sequencing batch reactors at short SRTs. Water Res 44:2141–2152. https://doi.org/10.1016/j.watres.2009.12.031
Yim SS, Choi JW, Lee YJ, Jeong KJ (2023) Rapid combinatorial rewiring of metabolic networks for enhanced poly(3-hydroxybutyrate) production in Corynebacterium glutamicum. Microb Cell Fact. https://doi.org/10.1186/s12934-023-02037-x
Wang J, Chen Z, Deng X et al (2023) Engineering Escherichia coli for Poly-β-hydroxybutyrate production from methanol. Bioengineering. https://doi.org/10.3390/bioengineering10040415
Author information
Authors and Affiliations
Contributions
BS involved in sourcing data and writing of manuscript. SB involved in sourcing data and writing of manuscript. RT involved in writing of manuscript. LMS involved in designing the manuscript, reviewing the manuscript, and sourcing of data.
Corresponding author
Ethics declarations
Conflict of interest
The authors of this manuscript have no affiliations or involvement in any entity or organization with any financial or non-financial interest in the subject discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Subramanian, B., Basak, S., Thirumurugan, R. et al. Metabolic flux analysis and metabolic engineering for polyhydroxybutyrate (PHB) production. Polym. Bull. (2024). https://doi.org/10.1007/s00289-024-05215-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00289-024-05215-y