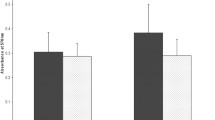
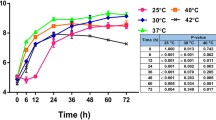
Abstract
In the present study, a comprehensive proteomic analysis of Brucella melitensis (B. melitensis) strain ATCC23457 was carried out to investigate proteome alterations in response to in vitro-induced nutrient stress. Our analysis resulted in the identification of 2440 proteins, including 365 hypothetical proteins and 850 potentially secretory proteins representing ~77.8% of the B. melitensis proteome. Utilizing a proteogenomics approach, we provide translational evidence for eight novel putative protein-coding genes and confirmed the coding potential of 31 putatively annotated pseudogenes, thus refining the existing genome annotation. Further, using a label-free quantitative proteomic approach, new insights into the cellular processes governed by nutrient stress, including enrichment of amino acid metabolism (E), transcription (K), energy production and conversion (C), and biogenesis (J) processes were obtained. Pathway analysis revealed the enrichment of survival and homeostasis maintenance pathways, including type IV secretion system, nitrogen metabolism, and urease pathways in response to nutrient limitation. To conclude, our analysis demonstrates the utility of in-depth proteomic analysis in enabling improved annotation of the B. melitensis genome. Further, our results indicate that B. melitensis undergoes metabolic adaptations during nutrient stress similar to other Brucella. sp, and adapts itself for long-term persistence and survival.





Similar content being viewed by others
References
Corbel MJ (1997) Brucellosis: an overview. Emerg Infect Dis 3(2):213
Majalija S, Luyombo P, Tumwine G (2018) Sero-prevalence and associated risk factors of Brucellosis among Malaria negative febrile out-patients in Wakiso district,Central Uganda. BMC Res Notes 11(1):1–6
Subbannayya Y, Pinto SM, Gowda H, Prasad TK (2016) Proteogenomics for understanding oncology: recent advances and future prospects. Expert Rev Proteom 13(3):297–308
Kucharova V, Wiker HG (2014) Proteogenomics in microbiology: taking the right turn at the junction of genomics and proteomics. Proteomics 14(23–24):2360–2675
Yang Y, Hu M, Yu K, Zeng X, Liu X (2015) Mass spectrometry-based proteomic approaches to study pathogenic bacteria-host interactions. Protein Cell 6(4):265–274
Mol JP, Pires SF, Chapeaurouge AD, Perales J, Santos RL, Andrade HM, Lage AP (2016) Proteomic profile of Brucella abortus-infected bovine chorioallantoic membrane explants. PLoS ONE 11(4):e0154209
Lauer SA, Iyer S, Sanchez T, Forst CV, Bowden B, Carlson K et al (2014) Proteomic analysis of detergent resistant membrane domains during early interaction of macrophages with rough and smooth Brucella melitensis. PLoS ONE 9(3):e91706
Connolly JP, Comerci D, Alefantis TG, Walz A, Quan M, Chafin R et al (2006) Proteomic analysis of Brucella abortus cell envelope and identification of immunogenic candidate proteins for vaccine development. Proteomics 6(13):3767–3780
Wagner MA, Eschenbrenner M, Horn TA, Kraycer JA, Mujer CV, Hagius S et al (2002) Global analysis of the Brucella melitensis proteome: identification of proteins expressed in laboratory-grown culture. Proteomics 2(8):1047–1060
Wang Y, Chen Z, Qiao F, Ying T, Yuan J, Zhong Z, Huang L (2009) Comparative proteomics analyses reveal the vir B of B. melitensis affects expression of intracellular survival related proteins. PLoS ONE 4(4):e5368
Pinto SM, Verma R, Advani J, Chatterjee O, Patil AH, Kapoor S et al (2018) Integrated multi-omic analysis of Mycobacterium tuberculosis H37Ra redefines virulence attributes. Front Microbiol 9:1314
Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J et al (2011) Global quantification of mammalian gene expression control. Nature 473(7347):337–342
Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278(5338):631–637
Bhatia VN, Perlman DH, Costello CE, McComb ME (2009) Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem 81(23):9819–9823
Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17(4):349–356
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Brinkman FS (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13):1608–1615
Walter W, Sánchez-Cabo F, Ricote M (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31(17):2912–2914
Prasad TK, Mohanty AK, Kumar M, Sreenivasamurthy SK, Dey G, Nirujogi RS et al (2017) Integrating transcriptomic and proteomic data for accurate assembly and annotation of genomes. Genome Res 27(1):133–144
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43(D1):D257–D260
Argyrou A, Vetting MW, Blanchard JS (2004) Characterization of a new member of the flavoprotein disulfide reductase family of enzymes from Mycobacterium tuberculosis. J Biol Chem 279(50):52694–52702
Gagic D, Ciric M, Wen WX, Ng F, Rakonjac J (2016) Exploring the secretomes of microbes and microbial communities using filamentous phage display. Front Microbiol 7:429
Lerat E, Ochman H (2005) Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res 33(10):3125–3132
Hong PC, Tsolis RM, Ficht TA (2000) Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun 68(7):4102–4107
Roop RM, Gaines JM, Anderson ES, Caswell CC, Martin DW (2009) Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198(4):221–238
Hidese R, Mihara H, Kurihara T, Esaki N (2012) Pseudomonas putida PydR, a RutR-like transcriptional regulator, represses the dihydropyrimidine dehydrogenase gene in the pyrimidine reductive catabolic pathway. J Biochem 152(4):341–346
Bao Y, Tian M, Li P, Liu J, Ding C, Yu S (2017) Characterization of Brucella abortus mutant strain Δ22915, a potential vaccine candidate. Vet Res 48(1):1–13
Singh AK, Balakrishna K, Sripathy MH, Batra HV (2014) Studies on recombinant glucokinase (r-glk) protein of Brucella abortus as a candidate vaccine molecule for brucellosis. Vaccine 32(43):5600–5606
Rest RF, Robertson DC (1974) Glucose transport in Brucella abortus. J Bacteriol 118(1):250–258
Vollmer W, Blanot D, De Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32(2):149–167
Wang Z, Bie P, Cheng J, Lu L, Cui B, Wu Q (2016) The ABC transporter YejABEF is required for resistance to antimicrobial peptides and the virulence of Brucella melitensis. Sci Rep 6(1):1–10
Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P et al (2005) Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect Immun 73(9):5578–5586
Delrue RM, Deschamps C, Léonard S, Nijskens C, Danese I, Schaus JM et al (2005) A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol 7(8):1151–1161
Weeks JN, Galindo CL, Drake KL, Adams GL, Garner HR, Ficht TA (2010) Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol 10(1):1–20
Dozot M, Poncet S, Nicolas C, Copin R, Bouraoui H, Mazé A et al (2010) Functional characterization of the incomplete phosphotransferase system (PTS) of the intracellular pathogen Brucella melitensis. PLoS ONE 5(9):e12679
Deutscher J, Francke C, Postma PW (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70(4):939–1031
Stülke J (2007) Regulation of virulence in Bacillus anthracis: the phosphotransferase system transmits the signals. Mol Microbiol 63(3):626–628
Sakaizawa T, Matsumura T, Fujii C, Hida S, Toishi M, Shiina T et al (2018) Potential role of ASC, a proapoptotic protein, for determining the cisplatin susceptibility of lung cancer cells. Tohoku J Exp Med 244(2):133–144
Wang Z, Wang S, Wu Q (2014) Cold shock protein A plays an important role in the stress adaptation and virulence of Brucella melitensis. FEMS Microbiol Lett 354(1):27–36
Jiang W, Hou Y, Inouye M (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem 272(1):196–202
Robertson GT, Roop RM (1999) The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol 34(4):690–700
Haine V, Dozot M, Dornand J, Letesson JJ, De Bolle X (2006) NnrA is required for full virulence and regulates several Brucella melitensis denitrification genes. J Bacteriol 188(4):1615–1619
Köhler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, Liautard JP (2002) The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci USA 99(24):15711–15716
Wang M, Qureshi N, Soeurt N, Splitter G (2001) High levels of nitric oxide production decrease early but increase late survival of Brucella abortus in macrophages. Microb Pathog 31(5):221–230
Porte F, Liautard JP, Köhler S (1999) Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun 67(8):4041–4047
Gajiwala KS, Burley SK (2000) HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol 295(3):605–612
Valderas MW, Alcantara RB, Baumgartner JE, Bellaire BH, Robertson GT, Ng WL et al (2005) Role of HdeA in acid resistance and virulence in Brucella abortus 2308. Vet Microbiol 107(3–4):307–312
Acknowledgements
We thank Karnataka Biotechnology and Information Technology Services (KBITS), Government of Karnataka, for support to the Center for Systems Biology and Molecular Medicine at Yenepoya (Deemed to be University) under the Biotechnology Skill Enhancement Programme (BiSEP) in Multiomics Technology. SMP is a recipient of INSPIRE Faculty Award from the Department of Science and Technology (DST), Government of India. SKB is a recipient of Bioinformatics National Certification (BINC)-Junior Research Fellowship from the Department of Biotechnology (DBT), Government of India. NA is a recipient of research fellowship assistance from YU. We thank Saketh Kapoor for his assistance in sample processing and data analysis.
Funding
This study was funded by Indian Council of Medical Research, Govt. of India [Grant No. Zon.15/11/2014-ECD-II].
Author information
Authors and Affiliations
Contributions
AAH, SMP, TSKP, HFD and RSK contributed to conception and design of the study. AAH, PRK, NMB, LRS, RSK provided samples for the study. SMP, and NA processed the samples. SMP, YS and SKB acquired and analyzed the data. AAH, SMP, YS and SKB prepared tables and figures. AAH, SMP, YS carried out the literature search and wrote sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical Approval
All protocols for sample collection from humans were approved by the Ethical Committee of the Dr. G.M. Taori Central India Institute of Medical Sciences (CIIMS).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Husain, A.A., Pinto, S.M., Agarwal, N. et al. Comprehensive Proteomic Analysis of Brucella melitensis ATCC23457 Strain Reveals Metabolic Adaptations in Response to Nutrient Stress. Curr Microbiol 80, 20 (2023). https://doi.org/10.1007/s00284-022-03105-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03105-y