Abstract
India was severely affected by several waves of SARS-CoV-2 infection that occurred during April–June 2021 (second wave) and December 2021–January 2022 (third wave) and thereafter, resulting in >10 million new infections and a significant number of deaths. Global Initiative on Sharing Avian Influenza Data database was used to collect the sequence information of ~10,000 SARS-CoV-2 patients from India and our sequence analysis identified three variants B.1.1.7 (alpha, α), B1.617.2 (delta, Δ), B.1.1.529 (Omicron, Oo) and one Omicron sub-variant BA.2.75 as the primary drivers for SARS-CoV-2 waves in India. Structural visualization and analysis of important mutations of alpha, delta, Omicron and its sub-variants of SARS-CoV-2 Receptor-Binding Domain (RBD) was performed and our analysis clearly shows that mutations occur throughout the RBD, including the RBD surface responsible for human angiotensin-converting enzyme 2 (hACE-2) receptor-binding. A comparison between alpha, delta and omicron variants/sub-variants reveals many omicron mutations in the hACE-2 binding site and several other mutations within 5 Å of this binding region. Further, computational analysis highlights the importance of electrostatic interactions in stabilizing RBD-hACE-2-binding, especially in the omicron variant. Our analysis explores the likely role of key alpha, delta and omicron mutations on binding with hACE-2. Taken together, our study provides novel structural insights into the implications of RBD mutations in alpha, delta and omicron and its sub-variants that were responsible for India’s SARS-CoV-2 surge.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
So far the SARS-CoV-2 global pandemic has caused >0.5 billion infections and >5 million deaths worldwide (www.worldometers.info/coronavirus). SARS-CoV-2 vaccines have proven to be effective but the threat of a rapidly evolving new virus variants and sub-variants that acquires multiple sets of new mutations requires continuous monitoring [1,2,3,4]. India underwent a surge of SARS-CoV-2 infections in early 2021 (second wave) and December 2021–January 2022 (third wave) and its threat still looms in India due to emergence of several sub-variants of omicron including the highly infectious BA.2.75 [5, 6]. The presence of emerging variants/sub-variants was speculated to be the primary driver of the infection burden.
A total of seven coronaviruses are known to infect humans [7]. While most are found to circulate and cause mild common cold-like symptoms, SARS-CoV and MERS-CoV were the only two coronaviruses of concern before SARS-CoV-2 was recognized [8, 9]. In SARS-CoV-2, membrane (M) and the Envelope (E) protein are involved in virus budding, the spike glycoprotein (S) is responsible for successful ingress of the virus [10,11,12]. The spike protein is a multi-domain protein that comprises of two subunits. The subunit S1 domain is mainly involved in the binding of the virus to the peptidase domain of the host surface receptor angiotensin-converting enzyme 2 (hACE-2), whereas subunit S2 is involved in viral attachment and entry [13,14,15]. On the mature virus, the spike protein exists as a trimer, with three receptor-binding domains (RBD) coming together that form the primary point of contact with hACE-2 and are the most dominant antigenic site [13, 14]. Thus, a large proportion of neutralizing antibody, vaccine, and therapeutic strategies target the RBD region [11, 16]. RBD is a hotspot of mutation as these mutations provide a fitness advantage to the virus either via direct impact on the binding interaction with hACE-2 receptor thus increasing infectivity or altering other aspects of virus biology such as pathogenicity or transmissibility [17]. Based on the site of mutation occurring in spike protein, SARS-CoV-2 is classified into several genomic variants [nomenclature is based on pango lineages (www.cov-lineages.org)]. Five variants of SARS-CoV-2 are major variants of concerns [(α), (β), (γ), (Δ) and (Oo)] as they are either highly transmissible, or more infective, or cause more severe disease than the original Wuhan-Hu-1 virus isolates [18].
Since RBD is a critical determinant of SARS-CoV-2 virus and host hACE-2 interaction, it is important to understand whether mutations in the RBD alter binding affinity to hACE-2. Such analysis will directly impact our understanding of the adaptive advantage towards virus infectivity, transmissibility, and viral immune evasion. Thus, this work aimed to analyse the available SARS-CoV-2 sequences from the second wave, third wave and afterwards in India and decipher the virus variant(s) sub-variants responsible for SARS-CoV-2 escalation in India. Our sequence analysis shows that SARS-CoV-2 alpha, delta and omicron variants were accountable for a vast majority of cases during the second and third wave in India [19]. We also found that different sub-variants of omicron including BA.2.75 emerged after third wave. We then structurally mapped key mutations onto the three-dimensional structures of RBD to comprehend their structural positions and assess their impact on RBD and hACE-2 interaction. We found several mutations throughout the RBD with many clustered together in patches of electrostatic-to-hydrophobic and/or hydrophobic to electrostatic. As a high number of RBD mutations are observed in the omicron and delta variants in and around the hACE-2-binding site, these may affect binding. Taken together, our analysis showcases several structural insights into the mutations within SARS-CoV-2 RBD of alpha, delta and omicron and their sub-variants in India.
Material and Methods
Data Collection
Full-length nucleotide sequences (total ~10,000 isolates) of SARS-CoV-2 deposited from India were collected from Global Initiative on Sharing All Influenza Database (GISAID) database (www.gisaid.org) between 31 May to June 15, 2021, December 2021 to January 2022 and May 2022 to August 2022. We set the selection criteria of viral genomes which exclusively infect human hosts and excluded those that carry low coverage and incomplete sequences [20, 21].
Sequence Alignment and Analysis
Emboss transeq (www.ebi.ac.uk/Tools/st/emboss_transeq/) was used to convert nucleotide sequence to protein [22]. Protein domains of SARS-CoV-2 was identified using Pfam (www.pfam.xfam.org), InterPro (www.ebi.ac.uk/interpro/), and SMART (www.smart.embl-heidelberg.de/) protein domain annotation server. Mutations present in different variants were identified by the aligning sequence of interests to the reference spike glycoprotein sequence (Wuhan-Hu-1, China uniprot ID: P0DTC2; GISAID ID: EPI_ISL_402124). ClustalW (www.genome.jp/tools-bin/clustalw) and Mafft (www.ebi.ac.uk/Tools/msa/mafft/) was used for multiple sequence alignments [22].
Structural Mapping
The three-dimensional structure of spike glycoprotein and hACE-2-RBD complex were accessed from Protein Data Bank (PDB IDs: 6LZG; www.rcsb.org) [23]. Interacting residues of hACE-2 and spike RBD were identified using PISA (www.ebi.ac.uk/pdbe/pisa/) and PDBsum (www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html). Variable regions of hACE-2 were identified using Consurf (https://consurf.tau.ac.il/). Non-synonymous mutations of hACE-2 were collected from dbSNP (www.ncbi.nlm.nih.gov/snp/) and these were processed using Ensembl (www.asia.ensembl.org/index.html). Charge and other physic chemical parameters were calculated using ProtParam (www.expasy.org/resources/protparam). Structure visualization, superimposition, and neighbouring residue identification were done using Pymol.
Computational Analysis of Variants
Computational calculations of solvent accessible surface area (SASA), thermal-stability and hydrophobicity (solubility) were done using modelled mutant structures of alpha, delta and omicron due to the unavailability of experimental three-dimensional structures. Modelling was done using Pymol followed by refinement using GalaxyWEB (https://galaxy.seoklab.org/) and Chiron (https://dokhlab.med.psu.edu/chiron/login.php). SASA was calculated using GetArea (http://curie.utmb.edu/getarea.html), thermo-stability was calculated using Scoop (http://babylone.ulb.ac.be) and hydrophobicity was calculated using SoDoPe (https://tisigner.com/sodope). HDOCK server (http://hdock.phys.hust.edu.cn/) was used for docking analysis between the RBD and hACE2 [23]. Binding affinity (ΔG) (kcal/mol) and dissociation constant (KD) (M) at 25 °C between RBD and receptor was determined using Prodigy website (https://wenmr.science.uu.nl/prodigy/). Infectivity is predicted based on Varsite (https://ebi.ac.uk/thornton-srv/databases/VarSite) analysis.
Results
Alpha (α), Delta (Δ) and Omicron (Oo) Variants were Found in Second and Third SARS-CoV-2 Wave in India
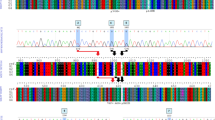
We collected the SARS-CoV-2 data from ~10,000 sequences from the GISAID database from 16 states and union territories of India. The state of Maharashtra accounted for the highest number of isolates (~30% of total samples) as the state has continued to report a high number of SARS-CoV-2 infections. Our sequence analysis detected more than 30 variants of SARS-CoV-2 across India; however, two variants (alpha variant) and (delta variant) were found in the majority of isolates especially after February 2021 (Fig. 1). The alpha variant was first acknowledged in the United Kingdom in late 2020 and carries more than 15 mutations in its spike protein. The delta variant, detected in India in October 2020 typically shows ~20 mutations in its spike protein [24]. SARS-CoV-2 data from across India revealed that the month of January had no sequences that corresponded to the delta variant, and 1% of cases in February corresponded to the delta variant (Fig. 1). Similarly, alpha variant cases were also low in these two months with only 5% in January. However, this number increased to 11% of the total sequences in February and other variants like B1.36; B1.1.216; B1.1 were also found dominant in the majority of the isolates (Fig. 1). Then, in March 2021 alpha variant was found in 22% sequenced samples followed by the delta variant (11%). However, we observed that this trend completely shifted in April 2021 and sequences from the delta variant dominated at 62% and by May, 94% of sequences were of the delta variant. On the other hand, sequences assigned to the alpha variant decreased to 11% in April and further to a minuscule 1% by May 2021 (Fig. 1). A similar trend was observed in Delhi with the alpha variant found in almost 41% of samples sequenced in February while the delta variant was virtually missing (Fig. 1). By March, while alpha variant cases were still observed at ~67%, cases attributed to delta variant exponentially increased with 64% of all sequences being of the delta variant by April and this continued to dominate (59%) in May (Fig. 1). Interestingly, during the third wave in India from December 2021 to January 2022, though 80% isolates sequences are of delta, however, 20% of isolates are of omicron (B.1.1529) and around 15–30 mutations are in the RBD in most of the isolates sequenced (Fig. 1). According to our sequence analysis of May up till August 2022, different sub-variants of the Omicron emerged all over the country, including Delhi. Among the different sub-variants of Omicron, most notable is B.A.2.75, which was found more than 50% of the sequenced sample in August 2022. BA.2.75 was first detected in the month of May 2022 and from June ends it had started to rise [25].
Trends of SARS-CoV-2 variants during the second and third outbreak of COVID-19 (January–May 2021 and December 2021–January 2022) in India and Delhi region. All sequence-related data was collected from GISAID (https://www.gisaid.org). The SARS-CoV-2 predominant variants found are B 1.1.7 (alpha), B1.617.2 (delta) and B.1.1.529 (omicron)
Unique Mutations are Present in the Variants
The full size of the SARS-CoV-2 genome is ~30 kb and spike protein is approximately ~3.9 kb (1273 aa) spanning ~10% of the total viral genome [26, 27]. The alpha and delta variants are characterized by few unique RBD mutations, e.g., the alpha variant is known for its characteristic N501Y and E484K, whereas L452R and T478K are the characteristic mutations associated with delta variant (Fig. 2A–B). These four RBD mutations are of interest as studies show a direct link between these mutations and SARS-CoV-2 infectivity, severity, and immune escape [28,29,30]. In contrast, the omicron variant exhibits a higher number of mutations in the RBD and around 15 mutations are predominately found in sequenced isolates (G339D, S371L, S373P, S375F, D405N, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H) (Fig. 2A–B) [31]. The major difference between original Omicron and its sub-variants BA.2.75 lies in three main mutations D339H, N460K, and R493Q in the RBD domain (Fig. S2). According to clinical data BA.2.75 is twenty to thirty percent more infectious compared to the base Omicron variant [32]. Interestingly, BA.2.75 emerged from BA.5 sub-variant of Omicron and in-general carries more mutations in the spike protein compared to the other sub-variants. Further, structural mapping revealed that E484K and N501Y in RBD of the alpha variant were located on the surface of a hairpin loop (E484) and in a loop region before the helix H7 (N501), respectively (Fig. 2A–B; Fig. S1). Another key mutant L452R reported in the delta variant was also found surface exposed and lay in a beta sheet (E) (Fig. 2B). Remarkably, many key omicron variant mutations mostly lie in the loop regions that constitute the hACE-2-binding site (G446S, S477N, T478K, E484A, G496S, Q498R, N501Y, and Y505H). However, few mutations are located in the core region in loops or beta sheet in the omicron variant. Previous investigation identifies immune escaping nature of G446S and Q493R mutation, and high abundance of both these mutation within BA.2.75 sub-variants probably provides an edge in bypassing immune checkpoints. Specifically, N460K and R493Q of this sub-variant are polar-to-charged and charged-to-polar mutations leading to likely change in electrostatic surface.
Key mutations in Spike Receptor Binding Domain (RBD) of B.1.1.7 (alpha variant), B.1.617.2 (delta variant) and B.1.1.529 (omicron) A Domain diagram of SARS-CoV-2 spike protein. RBD is coloured magenta and all domains other domains are coloured brown. The key RBD mutations in alpha, delta and omicron variants are listed in green B Structural mapping of key RBD mutations corresponding to the alpha, delta and omicron variants of SARS-CoV-2 (PDB ID: 7A98). RBD is coloured magenta and shown as surface. Mutant residues are represented as green spheres and marked (Color figure online)
Mutation Profile of RBD Domain
Structurally, RBD domain is composed of loop regions (more than 50%), and the core structure of RBD is formed by an antiparallel β sheet (twisted) which has small connecting helices and flexible loops (Fig. S1) [33]. The loop regions of the RBD are dynamic which imparts flexibility to residues particularly at the hACE-2 receptor-binding interface. Our analysis revealed that from beginning of the SARS-CoV-2 up to this point there was a continuous increase in the number of mutations occurring in the RBD domain, with G446S, N440K found common in alpha, delta, omicron and their sub-variants. This shows that such positions are highly flexible for mutation (Table S1). Furthermore, the continuous stretch of residues between 439 and 446 is found to be highly flexible accommodating several different types of mutations in the variants. Moreover, the region consisting of residues 450–500 is highly prone to mutation in alpha, delta and omicron variants (Table 1). Interestingly, in the omicron variant, both the N and C-termini of the RBD bear mutations (G339D, A520E, G526K, P527K) not seen before in alpha or delta variants in India. Interestingly, all these mutations are associated with altering charge of wild type RBD. Also, several mutations between residues 339–379 (S371L, S373P, S375F, T376A and C379R) are seen for the first time in India in the omicron variant (Table 1). These new mutations in the omicron variant are accompanied with either increase in hydrophobicity or positive surface charge. Also, between 400 and 450 residues, D405S mutation is seen in the omicron which accompanies charged-to-polar change on the surface.
Structural mapping of RBD mutations from alpha and delta variants (Table 1) revealed 8 non-conservative mutations (C488F, Q506K, P507L, S4771I, Y451H, G446V, T478K, E471G) that lie within 5 Å of hACE-2-binding residues in the delta variant (Fig. 3C–D). Of these, Q506K, T478K and Y451H are likely to affect the charge in the binding region. Also, Y451H mutation that lies in the proximity of L452R is likely to enhance the positively charged surface in this region. In contrast, in the alpha variant, only two mutations V445A (conserved hydrophobic) and G446V was seen in the proximity of hACE-2-binding region (Fig. 3A–B). Additionally, in the delta variant, 11 other mutations which were found to be distant from the hACE-2-binding site might play an additional role in the interaction (Fig. 3C–D). Of these, R346G, Q414R, G413V, A348S and A352S are located at the core central region of the RBD and are likely responsible for change in surface area and surface properties of the domain which may increase or restrict binding with hACE-2 receptor (Fig. 3D). Interestingly, the alpha variant had only one mutation in the core region (Fig. 3B). Finally, we identified three stretches on the delta variant where there is a possibility of several mutations clustering (Fig. S3). The first stretch is at the core of the RBD, the second stretch is around the notable mutation L452R and third stretch is around the second notable mutation T478K of the delta variant. Further, mutations G446V and A520S are seen in both alpha and delta variants. Our analysis revealed many other mutations apart from E484K, N501Y, L452R, and T478K (Table 1) which could be of possible significance especially when a large change in electrostatic surface is involved (Table S1, Fig. 3, Fig. S4). In contrast to alpha and delta, majority of mutations in omicron variant lie in the hACE-2-binding site (Fig. 3E, F). Further, 12 neutral residues mutate to charged residues and of these, 4 lie in the hACE-2-binding region (N417K, Q493R, Q498R, Y505H) having the ability to alter interaction with hACE-2 [34, 35]. Our analysis further shows 1% change in the surface area of hAC2-binding region due to mutations in the omicron variant (Table 2). Also, mutations E484A/Q, S477N and T478K are present within 5 Å of the main binding site of omicron variants of SARS-CoV-2 and these residues are found mutated in alpha and delta variants (Fig. 3).
Structural mapping of mutations observed in the RBD domain of B.1.1.7 (alpha variant), B.1.617.2 (delta variant) and B.1.1.529 (omicron variant) from India found in the GISAID database from January–May 2021 and December 2021–January 2022. A, C, E RBD mutations in alpha, delta and omicron variants. RBD is coloured violet and hACE-2 is coloured pale cyan. RBD-binding residues of hACE-2 are shown as sticks. The mutations which overlap with the binding region of hACE-2 are coloured red and shown as sticks. The mutations that lie within 5 Å of the hACE-2-binding residues of RBD are coloured green and shown as sticks. Other hACE-2-binding residues are coloured yellow. B, D, F RBD mutations in alpha, delta and omicron variants. RBD is coloured violet, hACE-2 is coloured pale cyan and rest of Spike is coloured brown and shown as a transparent surface. Mutations that exhibit a change in surface charge are shown as spheres and coloured blue and red, polar-to-hydrophobic mutations are coloured grey, hydrophobic to polar mutations are coloured cyan and remaining mutations are coloured orange. The key mutations are coloured green (Color figure online)
The Effects of RBD Mutations of Alpha, Delta and Omicron Variants on hACE-2 Binding
A total of 19 residues of RBD interact with hACE-2 in wild type structure of spike (Fig. S5). According to PDBSum analysis, the interaction between RBD and hACE-2 is formed by 11 h-bonds, 1 salt bridge and more than 100 non-bonding interactions. It is seen that the majority of the hACE-2-interacting residues are located in flexible loop regions of RBD. Our analysis shows that among the hACE-2-binding residues (K417, G446, Q493, G496, Q498, N501 and Y505), Q493 and Q498 mutate to arginine in the omicron variant not seen in the case of alpha and delta. This might facilitate stronger binding to negatively charged hACE-2 interface in the omicron variant [35]. Our hydrophobicity analysis of hACE-2 interface residues did not reveal any significant change in the solubility at the interface (Table 2). Further, the hACE-2 binding surface of RBD can be broadly divided into two patches [36, 37]. According to our analysis, patch 1 (Table 2, Fig. S6) is less conserved compared to patch 2, which is also smaller in size. Further, three out of four key RBD mutant residues are involved in direct interaction with hACE-2 in delta variant (Fig. S5). On the other hand, most of the mutations of the omicron are interestingly in patch 2 which was earlier mostly seen to be conserved (Fig. S6).
It is seen that wild type RBD N501 was associated with Y41 of hACE-2 via a non-bonding interaction; however, once this residue was mutated to tyrosine, this interaction increased several folds due to aromatic stacking (Fig. S7) [38]. The N501Y mutation is seen in both alpha and omicron variants. Mutations Y41R and Y41A in hACE-2 have been shown to increase binding affinity or abolish interaction, respectively. [39, 40] Another mutant residue E484 lies within 5 Å of hACE-2 K31 in most structures and the mutation E484K, seen in the alpha variant could stabilize charge at the binding surface forming an ion pair with hACE-2 E35 (Fig. S8). Further, hACE-2 mutations K31Y/D are shown in earlier studies to increase affinity or abolish interaction, respectively [40]. Interestingly, in comparison to alpha and delta, E484 mutates to either alanine or glutamine residue in the omicron variant which is likely to facilitate the binding by increasing hydrophobicity. Mutant residue L452, seen in both delta and omicron variants is the most intriguing as it does not directly interact with hACE-2, but its mutation L452R likely causes change in electrostatic association of RBD and hACE-2 (Fig. S9). Also, L452R could impact the interaction of neighbouring residues Q493, Y449 and Y453 with hACE-2 K31, E35, D38, and Q42 (Fig. S5–6). Interestingly, Q493 is found mutated to arginine in the omicron variant and a reversal happen in sub-variant BA.2.75. K417N mutation in the alpha, delta plus and omicron variants leads to a change in electrostatic properties and are likely to play a role in altered electrostatic interaction on the binding surface (Figs. S5, S9). Furthermore, superimposition of the modelled RBD variant structures of alpha, delta and omicron (PDB ID: 6LZG) exhibits high structural similarity (r.m.s.d. 0.3 Å) showing that hACE-2-binding interface of RBD in these variants are structurally conserved thereby suggesting that upto 15 mutations specifically in the omicron variant does not likely present a change in the overall stability of the RBD. Further, computational analysis of thermo-stability of variant structures verifies that omicron variant is little more stable compared to alpha and delta (Table 2). We further used docking to understand alpha, delta and omicron variants binding to hACE-2. Our docking result shows that BA.2.75 sub-variant of Omicron is likely to bind to hACE-2 with highest affinity (with topmost ΔG and docking score value), followed by Omicron, and alpha variant (Table S2). Reversion of R493Q is most likely responsible for enhanced affinity of the BA.2.75 RBD towards hACE-2 receptor. Also, our data clearly point out that the delta variant has a weaker affinity towards hACE-2 compared to wild type SARS-CoV-2; which is also in agreement with published reports. [23, 41]
hACE-2 Conservation and Crucial Residues for RBD-Binding
Structural analysis of hACE-2 has revealed that there are ~20 residues (mainly from the N-terminal domain of hACE-2) that are involved in interaction with RBD (Table 3) (Fig. S5). Out of twenty RBD-binding residues five residues are negatively charged, three residues are positively charged, three residue are hydrophobic and remaining nine residues are either aromatic/polar in nature. Due to the presence of high number of negative charge residues, hACE-2 binding interface is primarily negative. Since hACE-2 receptor is found abundant in almost all mammalian species, we studied hACE-2 protein conservation by performing a multiple sequence analysis (MSA) among several related mammalian species including humans, to understand the binding site conservation [42]. Our analysis showed that the RBD-binding site of hACE-2 was highly conserved which was in striking contrast to hACE-2 binding site of RBD, which we found to be highly evolved (Fig. S10). Since hACE-2 has the potential to affect SARS-CoV-2 infection, next we searched for the binding site and its adjacent residue mutation (variability) across different human alleles. Non-synonymous mutations present within different hACE-2 alleles were identified using established protocol [43]. We found several crucial mutations in critical RBD-binding residues in the human dbSNP database including T27A, E35D/K, E37K, M82I, P84T, and D355N (Table 4). Specifically, the charge-altering mutations E35K, E37K and D355N are found to be most essential in terms of altering electrostatic surface property and possibly decreasing the binding affinity with mostly positively charged RBD.
Discussion
Our analysis of SARS-CoV-2 sequences from second wave, third wave and afterword in India clearly shows dominance of the delta variant at the peak of the second wave (during early 2021), the occurrence of omicron variant in the third wave (December 2021–January 2022) and emergence of BA.2.75 sub-variant of Omicron in the current scenario. We observed that the mutational propensity of RBD alters with these variants with highest mutations observed for the omicron sub-variant BA.2.75. Both alpha and delta variants are known for their characteristic RBD mutations (N501Y and E484K for alpha and L452R and T478K for delta) and majority of these mutations were found in close proximity to hACE-2 binding interface. In contrast, a high number of 11 mutations in the omicron variant lie either at the hACE-2-binding site or within 5 Å. Our analysis shows that charge-altering mutations, especially positively charged, in the variants most likely can influence binding with the hACE-2 receptor as electrostatic interactions are seen to play a critical role in interaction between RBD and hACE-2 since hACE-2 binding surface is highly negative. On the other hand, hydrophobicity of the hACE-2 binding surface it seen to remain unchanged in the variants. Further analysis revealed key mutations in the RBD including N501Y seen in alpha and omicron variants and L452R seen in delta and omicron variants. N501Y mutation of RBD stabilizes hACE-2 binding through increased hydrophobic interaction with Y41 of hACE-2. On the contrary, L452R does not directly interact with hACE-2. As per our analysis leucine 452 mutation to arginine influences neighbouring residues (for e.g., Q493, Y453 and Y449). Interestingly, Q493R mutation is seen in the omicron variant. Further, the variants exhibit low structural variance with marginal higher thermo-stability in the omicron variant. Taken together, our structural analysis sheds insights into the mutations in alpha, delta, omicron, and its sub-variant BA.2.75 of SARS-CoV-2 that are responsible for dramatic increase in SARS-CoV-2 infections in India and their likely role in altering interaction with the hACE-2 receptor.
Conclusion
India witnessed a massive number of SARS-CoV-2 infections and considerable number of deaths during second third wave of infection, though the number of SARS-CoV-2 infection in last few months reduces significantly but it’s not completely eradicated from Indian population. In the current article, we have explored the status of SARS-CoV-2 variants and sub-variants during the second/third wave and afterwards in India. Subsequently we mapped all important mutations onto the three-dimensional structure of receptor-binding domain (RBD) of spike protein to understand the impact of mutation on receptor-binding. Furthermore, we found that a substantial number of RBD residues exhibit structural plasticity due to mutation, accompanied by a change in surface electrostatic. Our analysis further predicts that electrostatic variation of RBD also facilitates extended interaction with human-Angiotensin-Converting Enzyme 2 (hACE-2). According to our knowledge, this is the first comprehensive study showcasing structural details of alpha, delta, omicron and its sub-variants that emerged during last two years of SARS-CoV-2 infection in India.
Supporting Information Available
Supporting information contains different bioinformatics analysis related to the RBD region of spike protein and hACE-2 receptor and freely available online.
Data Availability
Individual GISAID sequences and other materials are available from the corresponding author upon request.
References
Williams TC, Burgers WA (2021) SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir Med 9(4):333–335. https://doi.org/10.1016/S2213-2600(21)00075-8
Gupta RK (2021) Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat Rev Immunol 21(6):340–341. https://doi.org/10.1038/s41577-021-00556-5
Yadav PD, Sapkal GN, Sahay RR, Potdar VA, Deshpande GR, Patil DY et al (2022) Substantial immune response in Omicron infected breakthrough and unvaccinated individuals against SARS-CoV-2 variants of concerns. J Infect 84(5):e80–e81. https://doi.org/10.1016/j.jinf.2022.02.005
Samarasekera U (2021) India grapples with second wave of COVID-19. Lancet Microbe 2:e238. https://doi.org/10.1016/S2666-5247(21)00123-3
Desingu PA, Nagarajan K (2022) SARS-CoV-2 Omicron variant is spreading in different parts of the world in three different trends. J Med Virol 94(6):2354–2356. https://doi.org/10.1002/jmv.27646
Jain VK, Iyengar KP, Vaishya R (2021) Differences between First wave and Second wave of COVID-19 in India. Diabetes Metab Syndr 15(3):1047–1048. https://doi.org/10.1016/j.dsx.2021.05.009
Chen B, Tian E-K, He B, Tian L, Han R, Wang S et al (2020) Overview of lethal human coronaviruses. Signal Transduct Target Ther 5(1):89. https://doi.org/10.1038/s41392-020-0190-2
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020) The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. https://doi.org/10.1038/s41564-020-0695-z
Hu B, Guo H, Zhou P, Shi Z-L (2021) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. https://doi.org/10.1038/s41579-020-00459-7
Huang Y, Yang C, Xu X, Liu S-W (2020) Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 41(9):1141–1149. https://doi.org/10.1038/s41401-020-0485-4
Wang MY, Zhao R, Gao L-J, Gao X-F, Wang D-P, Cao J-M (2020) SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol 10:587269. https://doi.org/10.3389/fcimb.2020.587269
Watanabe Y, Allen JD, Wrapp D, McLellan J-S, Crispin M (2020) Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369(6501):330–333. https://doi.org/10.1126/science.abb9983
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S et al (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE-2 receptor. Nature 581(7807):215–220. https://doi.org/10.1038/s41586-020-2180-5
Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM et al (2020) Distinct conformational states of SARS-CoV-2 spike protein. Science 369(6511):1586–1592. https://doi.org/10.1126/science.abd4251
Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C et al (2020) Molecular architecture of the SARS-CoV-2 virus. Cell 183(3):730-738.e13. https://doi.org/10.1016/j.cell.2020.09.018
Sadarangani M, Marchant A, Kollmann TR (2021) Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat Rev Immunol 21(8):475–484. https://doi.org/10.1038/s41577-021-00578-z
Li C, Tian X, Jia X, Wan J, Lu L, Jiang S et al (2021) The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Transduct Target Ther 6(1):132. https://doi.org/10.1038/s41392-021-00536-0
Kim S, Liu Y, Ziarnik M, Cao Y, Zhang F, Im W (2022) Binding of human ACE2 and RBD of Omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern. bioRxiv. https://doi.org/10.1101/2022.01.24.477633
Mahapatra RK, Tiwari R, Sarangi AK, Sharma SK, Khandia R, Saikumar G et al (2022) Twin combination of Omicron and Delta variants triggering a tsunami wave of ever high surges in COVID-19 cases: a challenging global threat with a special focus on the Indian subcontinent. J Med Virol 94(5):1761–1765. https://doi.org/10.1002/jmv.27585
Alai S, Gujar N, Joshi M, Gautam M, Gairola S (2021) Pan-India novel coronavirus SARS-CoV-2 genomics and global diversity analysis in spike protein. Heliyon 7(3):e06564. https://doi.org/10.1016/j.heliyon.2021.e06564
Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M (2021) SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 9(7):1542. https://doi.org/10.3390/microorganisms9071542
Kumar S, Karuppanan K, Subramaniam G (2022) Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. J Med Virol 94(10):4780–4791. https://doi.org/10.1002/jmv.27927
Vardhan S, Sahoo SK (2022) Computational studies on the interaction of SARS-CoV-2 Omicron SGp RBD with human receptor ACE2, limonin and glycyrrhizic acid. Comput Biol Med 144:105367. https://doi.org/10.1016/j.compbiomed.2022.105367
Rahbar MR, Jahangiri A, Khalili S, Zarei M, Zeinabad KM, Khalesi B et al (2021) Hotspots for mutations in the SARS-CoV-2 spike glycoprotein: a correspondence analysis. Sci Rep 11(1):23622. https://doi.org/10.1038/s41598-021-01655-y
Callaway E (2022) Will “Centaurus” be the next global coronavirus variant? Indian cases offer clues. Nature 608(7923):462–463. https://doi.org/10.1038/d41586-022-02154-4
Yang P, Wang X (2020) COVID-19: a new challenge for human beings. Cell Mol Immunol 17(5):555–557. https://doi.org/10.1038/s41423-020-0407-x
Bangaru S, Ozorowski G, Turner HL, Antanasijevic A, Huang D, Wang X et al (2020) Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science 370(6520):1089–1094. https://doi.org/10.1126/science.abe1502
Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C et al (2020) The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182(5):1284-1294.e9. https://doi.org/10.1016/j.cell.2020.07.012
Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EC et al (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7):409–424. https://doi.org/10.1038/s41579-021-00573-0
Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnani SE (2021) SARS-CoV-2 variants of concern are emerging in India. Nat Med 27(7):1131–1133. https://doi.org/10.1038/s41591-021-01397-4
Das S, Samanta S, Banerjee J, Pal A, Giri B, Kar SS et al (2022) Is Omicron the end of pandemic or start of a new innings? Travel Med Infect Dis 48:102332. https://doi.org/10.1016/j.tmaid.2022.102332
Tan WC, Lim B-L, Young BE, Yeoh AY-Y, Yung C-F, Yap W-C et al (2022) Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet Microbe. https://doi.org/10.1016/S2666-5247(22)00220-8
Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S et al (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 17(6):613–620. https://doi.org/10.1038/s41423-020-0400-4
Koley T, Kumar M, Goswami A, Ethayathulla AS, Hariprasad G (2022) Structural modeling of Omicron spike protein and its complex with human ACE-2 receptor: molecular basis for high transmissibility of the virus. Biochem Biophys Res Commun 592:51–53. https://doi.org/10.1016/j.bbrc.2021.12.082
Rath SL, Padhi AK, Mandal N (2022) Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem Biophys Res Commun 592:18–23. https://doi.org/10.1016/j.bbrc.2022.01.006
Liu K, Tan S, Niu S, Wang J, Wu L, Sun H et al (2021) Cross-species recognition of SARS-CoV-2 to bat ACE-2. Proc Natl Acad Sci U S A 118(1):e2020216118. https://doi.org/10.1073/pnas.2020216118
Niu S, Wang J, Bai B, Wu L, Zheng A, Chen Q et al (2021) Molecular basis of cross-species ACE-2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J 40(16):e107786. https://doi.org/10.15252/embj.2021107786
Zhu X, Mannar D, Srivastava SS, Berezuk AM, Demers JP (2021) Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE-2 and 2 potent neutralizing antibodies. PLoS Biol 19(4):e3001237. https://doi.org/10.1371/journal.pbio.3001237
Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM et al (2020) Engineering human ACE-2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369(6508):1261–1265. https://doi.org/10.1126/science.abc0870
Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S et al (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE-2. EMBO J 24(8):1634–1643. https://doi.org/10.1038/sj.emboj.7600640
Durmaz V, Kochl K, Krassnigg A, Parigger L, Hetmann M, Singh A et al (2022) Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference. Sci Rep 12(1):14534. https://doi.org/10.1038/s41598-022-18507-y
Bibiana SOF, Vargas-Pinilla P, Amorim CEG, Sortica VA, Bortolini MC et al (2020) ACE-2 diversity in placental mammals reveals the evolutionary strategy of SARS-CoV-2. Genet Mol Biol 43(2):e20200104. https://doi.org/10.1590/1678-4685-gmb-2020-0104
Hussain M, Jabeen N, Raza F, Shabbir S, Baig AA, Amanullah A (2020) Structural variations in human ACE-2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol 92(9):1580–1586. https://doi.org/10.1002/jmv.25832
Acknowledgements
We are thankful to NIMR, New Delhi for providing us necessary research infrastructure.
Funding
SC is supported by Ramalingaswami Fellowship (BT/RLF/Re-entry/09/2019) by DBT. AS is supported by the Department of Science and Technology (DST) JC Bose fellowship (JCB-41). RG is the recipient of a Junior research fellowship from the Dept of Biotechnology (DBTHRDPMV/JRF/BET-20/I/2020/AL/206), Govt of India. RG is also supported by AcSIR for PhD (Registration No. 10BB21J65012). SK is supported by the DBT/Wellcome Trust India Alliance Early Career Fellowship grant IA/E/18/1/504307.
Author information
Authors and Affiliations
Contributions
All authors contributed towards implementation of the research. AS designed the study. SC, JG, AS and RG collected and analysed the data. SC, JG, AS, AC, SK wrote the manuscript. All authors read and approved the final form of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chakraborti, S., Gill, J., Goswami, R. et al. Structural Profiles of SARS-CoV-2 Variants in India. Curr Microbiol 80, 1 (2023). https://doi.org/10.1007/s00284-022-03094-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03094-y