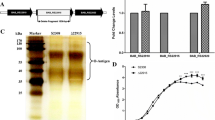
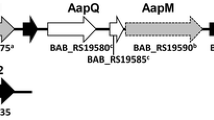
Abstract
It is widely acknowledged that pseudogenes play important roles in bacterial diversification and evolution and participate in gene regulation and RNA interference (RNAi). However, the function of most pseudogenes in Brucella spp remains poorly understood, warranting further studies.To comprehensively analyze the function of the pseudogenes BMEA_B0173 in Brucella melitensis strain 63/9, a BMEA_B0173 in-frame deleted mutant strain was constructed. Then, the phenotypes of the mutant strain, such as growth characteristics and bacterial virulence, were assessed in mice infection models. Finally, iTRAQ analysis was performed to investigate the gene expression profile affected by the pseudogenes BMEA_B0173. In this study, we found that BMEA_B0173 deletion exhibited increased agglutination with M monospecific sera. In a mouse model of chronic infection, the BMEA_B0173 deletion strain displayed increased colonization in the spleen compared to the wild-type pathogen. The iTRAQ assay revealed that 252 proteins were differentially expressed between the BMEA_B0173 deletion and the wild-type strains. In addition, deletion of BMEA_B0173 significantly increased the expression of proteins involved in the denitrification pathway, iron metabolism, and several transcriptional regulators, which might cause increased virulence of the mutant strain. In conclusion, this study preliminary uncovered the function of the pseudogene BMEA_B0173 in Brucella melitensis 63/9 and provided novel insights for studying the pathogenesis of Brucella strains.




Similar content being viewed by others
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD036210.
References
Ahmed W, Zheng K, Liu Z (2016) Establishment of chronic infection: Brucella’s stealth strategy. Front Cell Infect Mi 6:30. https://doi.org/10.3389/fcimb.2016.00030
Corbel M (1997) Brucellosis: an overview. Emerg Infect Dis 3(2):213–221. https://doi.org/10.3201/eid0302.970219
Suárez-Esquivel M, Chaves-Olarte E, Moreno E, Guzmán-Verri C (2020) Brucella genomics: macro and micro evolution. Int J Mol Sci 21(20):7749. https://doi.org/10.3390/ijms21207749
Wattam AR, Williams KP, Snyder EE, Almeida NF, Shukla M, Dickerman AW, Crasta OR, Kenyon R, Lu J, Shallom JM (2009) Analysis of ten brucella genomes reveals evidence for horizontal gene transfer despite a preferred intracellular lifestyle. J Bacteriol 191:3569
Suárez-Esquivel M, Baker KS, Ruiz-Villalobos N, Hernández-Mora G, Barquero-Calvo E, González-Barrientos R, Castillo-Zeledón A, Jiménez-Rojas C, Chacón-Díaz C, Cloeckaert A, Chaves-Olarte E, Thomson NR, Moreno E, Guzmán-Verri C (2017) Brucella genetic variability in wildlife marine mammals populations relates to host preference and ocean distribution. Genome Biol Evol 9(7):1901–1912. https://doi.org/10.1093/gbe/evx137
Dagan T, Blekhman R, Graur AD (2006) The “domino theory” of gene death: Gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol Biol Evol 23(2):310–316
Goodhead I, Darby AC (2015) Taking the pseudo out of pseudogenes. Curr Opin Microbiol 23:102–109. https://doi.org/10.1016/j.mib.2014.11.012
Sasidharan R, Gerstein M (2008) Protein fossils live on as RNA. Nature 453(7196):729–731. https://doi.org/10.1038/453729a
Balakirev ES, Ayala FJ (2003) Pseudogenes: are they “Junk” or functional DNA? Annu Rev Genet 37(1):123–151
Liu Y, Dong H, Peng X, Gao Q, Jiang H, Xu G, Qin Y, Niu J, Sun S, Li P, Ding J, Chen R (2019) RNA-seq reveals the critical role of Lon protease in stress response and Brucella virulence. Microb Pathogenesis 130:112–119. https://doi.org/10.1016/j.micpath.2019.01.010
Stranahan LW, Khalaf OH, Garcia-Gonzalez DG, Arenas-Gamboa AM (2019) Characterization of Brucella canis infection in mice. PLoS ONE 14(6):e218809. https://doi.org/10.1371/journal.pone.0218809
Varesio LM, Willett JW, Fiebig A, Crosson S (2019) A carbonic anhydrase pseudogene sensitizes select brucella lineages to low CO2 tension. J Bacteriol 201(22):e509–e519
Bialer MG, Ferrero MC, Delpino MV, Ruiz-Ranwez V, Zorreguieta A (2021) Adhesive functions or pseudogenization of type va autotransporters in brucella species. Front Cell Infect Mi 11:607610
Zygmunt MS, Blasco JM, Letesson J, Cloeckaert A, Moriyon I (2009) DNA polymorphism analysis of Brucella lipopolysaccharide genes reveals marked differences in O-polysaccharide biosynthetic genes between smooth and rough Brucella species and novel species-specific markers. Bmc Microbiol 9(1):92. https://doi.org/10.1186/1471-2180-9-92
Vemulapalli R, He Y, Buccolo LS, Boyle SM, Sriranganathan N, Schurig GG (2000) Complementation of Brucella abortus RB51 with a functional wboA gene results in O-Antigen synthesis and enhanced vaccine efficacy but no change in rough phenotype and attenuation. Infect Immun 68(7):3927–3932. https://doi.org/10.1128/IAI.68.7.3927-3932.2000
Wang X, Wang Y, Ma L, Zhang R, De Y, Yang X, Wang C, Wu Q (2015) Development of an improved competitive ELISA based on a monoclonal antibody against lipopolysaccharide for the detection of bovine brucellosis. Bmc Vet Res 11(1):118. https://doi.org/10.1186/s12917-015-0436-3
Stranahan LW, Arenas-Gamboa AM (2021) When the going gets rough: the significance of brucella lipopolysaccharide phenotype in host–pathogen interactions. Front Microbiol 12:713157. https://doi.org/10.3389/fmicb.2021.713157
Lapaque N, Moriyon I, Moreno E, Gorvel J (2005) Brucella lipopolysaccharide acts as a virulence factor. Curr Opin Microbiol 8(1):60–66. https://doi.org/10.1016/j.mib.2004.12.003
Fernandez-Prada CM, Zelazowska EB, Nikolich M, Hadfield TL, Roop RM, Robertson GL, Hoover DL (2003) Interactions between Brucella melitensis and human phagocytes: bacterial surface O-Polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect Immun 71(4):2110–2119. https://doi.org/10.1128/IAI.71.4.2110-2119.2003
Gross A, Spiesser S, Terraza A, Rouot B, Caron E, Dornand J (1998) Expression and bactericidal activity of nitric oxide synthase in Brucella suis -infected murine macrophages. Infect Immun 66(4):1309–1316. https://doi.org/10.1128/IAI.66.4.1309-1316.1998
Loisel-Meyer S, Jimenez D, Basseres E, Dornand J, Kohler S, Liautard JP, Jubier-Maurin V (2006) Requirement of norD for brucella suis virulence in a murine model of in vitro and in vivo infection. Infect Immun 74(3):1973–1976
Roop RM (2012) Metal acquisition and virulence in Brucella. Anim Health Res Rev 13(1):10–20. https://doi.org/10.1017/S1466252312000047
Paulley JT, Anderson ES, Roop RM (2007) Brucella abortus requires the heme transporter BhuA for maintenance of chronic infection in BALB/c mice. Infect Immun 75(11):5248–5254. https://doi.org/10.1128/IAI.00460-07
Bellaire BH, Elzer PH, Hagius S, Walker J, Baldwin CL, Roop RM (2003) Genetic organization and Iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect Immun 71(4):1794–1803. https://doi.org/10.1128/IAI.71.4.1794-1803.2003
Elhassanny AEM, Anderson ES, Menscher EA, Roop RM (2013) The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol Microbiol 88(6):1070–1082. https://doi.org/10.1111/mmi.12242
Almirón MA, Ugalde RA (2010) Iron homeostasis in Brucella abortus: the role of bacterioferritin. J Microbiol 48(5):668–673. https://doi.org/10.1007/s12275-010-0145-3
Xiong X, Li B, Zhou Z, Gu G, Li M, Liu J, Jiao H (2021) The VirB system plays a crucial role in brucella intracellular infection. Int J Mol Sci 22(24):13637. https://doi.org/10.3390/ijms222413637
Sun YH, Rolan HG, Hartigh AD, Sondervan D, Tsolis RM (2005) Brucella abortus VirB12 is expressed during infection but is not an essential component of the type IV secretion system. Infect Immun 73(9):6048–6054
Acknowledgements
We thank Shanghai Personal Biotechnology Cp. Ltd. for their support for bioinformatics analysis with their Analysis Platform.
Funding
This work was supported by the National Key Research and Development Program of China (NO.2016YFD0500902) and Natural Science Foundation of China(NO.31902310).
Author information
Authors and Affiliations
Contributions
JD and XS contributed to the study conception and design. Material preparation, data collection, and analyses were performed by GZ, YF, HJ, TW, XW, XP, YZ, XZ, and LZ. The first draft of the manuscript was written by GZ, JS, ML, and HD, and all the authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest, financial or otherwise.
Ethical Approval
This study was approved by the Animal Ethics Committee of the China Institute of Veterinary Drug Control.
Consent for Publication
Not applicable.
Research Involving in Animal Participants
Mice experiments were strictly performed in accordance with the Experimental Animal Regulation Ordinances set by the China National Science and Technology Commission.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2022_3078_MOESM2_ESM.tif
Supplementary file2 (TIF 3534 kb)—Figure S2. The enriched pathways of 10 genes (different colors represented different pathways)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, G., Dong, H., Feng, Y. et al. The Pseudogene BMEA_B0173 Deficiency in Brucella melitensis Contributes to M-epitope Formation and Potentiates Virulence in a Mice Infection Model. Curr Microbiol 79, 378 (2022). https://doi.org/10.1007/s00284-022-03078-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03078-y