Abstract
Background
Since recognizing the interaction between Brucella and host cells is crucial to the elucidation of the infectious process, Brucella researches have prioritized the investigation of genes related to pathogenicity. To demonstrate the roles of Brucella genes, RAW 264.7 cells were infected with the Brucella abortus wild-type and mutant strains (generated using transposon mutagenesis), after which the different transcriptional responses of the infected cells were determined using microarray.
Results
Following infection, enhanced strategies for intracellular survival, such as down-regulation of genes associated with cytokine responses and apoptosis, were observed in RAW 264.7 cells infected with C3 mutant strain when compared to the transcriptional responses of wild-type infected cells. Using sequence analysis, we determined the mutation site of a C3 mutant strain as the ATP-binding cassette transporter permease (BruAb2_1031). These results were evidenced by an increased level of intracellular survival of the C3 mutant strain.
Conclusions
Characteristics of each mutant strain including bacterial growth rate, abilities to induce cytokine production in macrophages after infection, internalization, and levels of intracellular survival and replication, were investigated by performing RAW 264.7 cell infection experiments. Our results indicate that the BruAb2_1031 gene might be closely related with intracellular survival of B. abortus in RAW 264.7 cells.
Similar content being viewed by others
Background
Brucella abortus (B. abortus), a member of the Alphaproteobacteria family, is a facultative intracellular bacteria that causes undulant fever, arthritis, endocarditis, and osteomyelitis in humans and abortion and infertility in cattle [1]. Unlike other bacterial pathogens, Brucella do not produce classical virulence factors such as exotoxins, cytolysins, capsules, fimbria, plasmids, lysogenic phage, and endotoxic lipopolysaccharide (LPS) molecules [1,2,3]. Rather, they invade and replicate in professional and non-professional phagocytic cells, thereby eluding the bactericidal immune responses of the host [4].
As facultative intracellular bacteria, Brucella can survive and replicate within host macrophages [1, 5]. To establish successful strategies for intracellular survival, Brucella often inhibit the normal functions of the host [6,7,8]. Indeed, Brucella utilizes macrophages for infection through mechanisms such as inhibition of apoptosis [6], modification of membrane-bound vesicles [7], and interruption of phagosome-lysosome formation [8]. Hence, identifying the interactions between Brucella and macrophages is a key role in understanding its pathogenesis [9, 10]. However, the pathogenic mechanisms of Brucella based on these interactions are not well understood.
In Brucella infection, macrophages play a central role as the first line of defense of the immune system and the primary target of the pathogen [11, 12]. Brucella infected macrophages are activated to promote killing of the bacteria by induction of superoxide anion and hydrogen peroxide [13]. Moreover, the infected macrophages produce pro-inflammatory cytokines (TNF-α, IL-6, and IL-12) and chemokines (GRO- α, IL-8, MCP-1, RANTES, and MIP1 α/β) as critical coordinators of adaptive immunity [5, 14]. Among these biological mediators, TNF-α strongly enhances the bactericidal activity of phagocytes, while IL-12 induces IFN-γ, the major cytokine fighting Brucella infection [5, 11]. Along with the cytokine activities, macrophages further respond to Brucella infection by inducing apoptosis, thereby exposing the bacterium to the extracellular environment and reducing its replication [6, 14, 15]. Reports indicate that these protective immune responses against Brucella are probably associated with several Brucella genes which are involved in components or functions of the bacteria, such as lipopolysaccharides, outer membrane proteins, heat shock proteins, ABC-type transporters, and Cu-Zn superoxide dismutase [16, 17]. However, Brucella genes associated with the host immune responses and bacterial survival need to be determined more clearly to control brucellosis based on its pathogenesis.
In this study, transcriptional responses of the mouse macrophage cell line (RAW 264.7) infected with B. abortus mutant strains were analyzed to demonstrate the role of Brucella genes. B. abortus mutant strains were previously generated using transposon mutagenesis [18], which is frequently used as a genetic tool to characterize genes of unknown function [19]. Through PCR and alignment analysis, the mutated genes of select mutant strains were revealed as ATP-binding cassette (ABC) transporter permease (BruAb2_1031), ABC transporter substrate-binding protein (BruAb2_0113), and alkyl hydroperoxide reductase D (ahpD). As reported previously, BruAb2_1031 and BruAb2_0113 have a role of importing peptides and iron into the bacteria as the ABC transporter system [20]. The ahpD is a peroxiredoxin reductase that restores the enzymatic activity of ahpC, thereby having an important role as an antioxidants [21]. However, the functional role of these genes is unclear in brucellosis. The transcriptional responses were determined by the microarray approach, which allows understanding of global cell responses. The roles of Brucella genes affecting the interactions between host immune cells and the bacteria have been discussed based on differences in the transcriptional responses between macrophages infected with the B. abortus mutant strains and the B. abortus wild-type.
Results
Characteristics of B. abortus mutant strains
Among the generated mutant strains, different characteristics were observed in the bacterial growth rates and product levels of NO, IL-6, and TNF-α in RAW 264.7 cells responding to infections of C3, C24 and C30 strains (Additional file 1: Table S1, Additional file 2: Figure S1, and Table 1). The mutant strains were divided into three groups according to the growth rate compared to the wild-type (Additional file 1: Table S1). The C3 mutant strain was selected in group A (mutants showing more than 10% reduction in growth rate) since cells infected by this strain showed lower product levels of NO, IL-6, and TNF-α as compared to others strains in the group. However, RAW 264.7 cells infected with strain C24 in group B (mutants showing similar growth rate) and strain C30 in group C (mutants showing more than 10% increment in growth rate) induced higher levels of NO and TNF-α, and lower levels of IL-6, as compared to other strains in their respective groups. NO and TNF-α are mainly associated with inflammatory reactions, but IL-6 contributes to both pro- and anti-inflammatory responses; hence, the mutant strains C24 and C30 were selected for further studies due to their unique characteristics as compared to other mutants in their group.
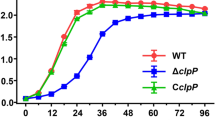
The levels of internalization, intracellular survival after internalization, and intracellular replication of each Brucella strain in RAW 264.7 cells were investigated (Fig. 1 and Additional file 3: Figure S2). Although the highest internalization level was observed in B. abortus wild-type (Fig. 1a, p < 0.05 and p < 0.01), the decrease was steeper than that observed in of the mutant strains, between 0 h and 6 h (Fig. 1b and c, p < 0.05). This steep decrease demonstrated that the wild-type has a lower level of intracellular survival as compared to the mutant strains. According to the CFU slope level increase between 12 h and 48 h, lower levels of intracellular replication were observed in the C3 and C30 mutant strains as compared to the wild-type (Fig. 1c, p < 0.01).
The levels of intracellular B. abortus wild-type and mutant strains in infected RAW 264.7 cells. RAW 264.7 cells were infected with wild-type and each mutant strain for 1 h at MOI 100, after which a gentamicin protection assay was conducted. At selected time points, the medium was removed and cells were washed prior to lysis, after which the lysate was plated on to brucella agar. a Internalization levels (0 h) of B. abortus wild-type and mutant strains in RAW 264.7 cells after gentamicin protection assay. b Intracellular CFU level at 6 h after internalization, indicating the levels of intracellular survival. c Intracellular CFU level of each strain at selected time points after internalization, indicating the levels of intracellular survival (6 h) and replication (12 h, 24 h, and 48 h) after internalization in RAW 264.7 cells. The CFU levels of (b) and (c) were expressed as a percentage compared to the respective CFU of internalization of each strain. (*p < 0.05 and **p < 0.01)
Identification of the transposon insertion site in B. abortus mutant strains
The insertion sequence of B. abortus mutant C3 was located at the 1,033,916 bp - 1,034,502 bp of chromosome II, B. abortus mutant C24 was at the 113,672 bp - 114,206 bp, and B. abortus mutant C30 was at the 529,584 bp - 530,364 bp region. From sequencing results, ABC transporter permease (BruAb2_1031), ABC transporter substrate-binding protein (BruAb2_0113) and alkyl hydroperoxide reductase D (ahpD) were identified as the genes disrupted by transposon insertion in mutant strains C3, C24, and C30, respectively.
Determination of differentially expressed genes of infected RAW 264.7 cells
Following infection of B. abortus wild-type for 6 h, 12 h, and 24 h, we observed more than 2-fold change in the expression of 58, 202, and 1766 genes, respectively, as compared to uninfected cells (Additional file 4: Table S2). The mutant strain infected RAW 264.7 cells also showed different gene expression levels when compared to uninfected cells (Additional file 5: Figure S3 and Additional file 6: Figure S4). The change in expression levels at 6 h, 12 h, and 24 h, respectively, were observed in: 48, 157, and 1239 genes in C3 mutant strain infected cells; 25, 164, and 1422 genes in C24 mutant strain infected cells; and 58, 202, and 1766 genes in C30 mutant strain infected cells.
In comparison to the wild-type infected RAW 264.7 cells, the C3 mutant strain infected cells showed 7, 9, and 57 altered genes at 6 h, 12 h, and 24 h, respectively (Fig. 2 and Additional file 7: Figure S5), whereas C24 and C30 infections showed number of altered genes less than 20 at all-time points (Fig. 2 and Additional file 7: Figure S5). Additional file 7: Figure S5 shows the median of normalized hybridization signals of genes with altered transcription comparing the wild-type and mutant strain infected RAW 264.7 cells at each time point. When compared to wild-type infected RAW 264.7 cells, all genes showing altered expression level of > |2| in B. abortus mutant strains infected cells are presented in Additional file 8 Table S3, Additional file 9 Table S4, and Additional file 10 Table S5.
Gene set enrichment analysis of infected RAW 264.7 cells
The genes showing altered expression were categorized by gene set enrichment analysis using the Protein Analysis Through Evolutionary Relationships (PANTHER) classification database, to demonstrate coordinated changes in pre-specified sets of related genes. Genes showing different regulation in mutant-infected RAW 264.7cells compared to wild-type infected cells were categorized by molecular functions and biological processes, as shown in Figs. 3 and 4, respectively. Most altered genes in C3 mutant strain infected cells for 24 h were down-regulated, compared to the wild-type infections. These regulated genes were mostly involved in the molecular functions of binding (Fig. 3b) and biological processes of cellular process, response to stimulus, and biological regulation (Fig. 4b). Also, most biological processes of cells were down-regulated in C3 mutant-infected RAW 264.7cells, while only few genes were-upregulated in the C24 mutant-infected cells (Fig. 4a). In case of C24 and C30 mutant strain infected cells, no significant results for pre-specified sets in the PANTHER classification system were observed.
Categorization by molecular function of genes, showing different expression levels between B. abortus infections. The different expression levels in B. abortus mutant strain infected RAW 264.7 cells were compared to the wild-type infected cells at 24 h after infection. a Up-regulated genes. b Down regulated genes
Categorization by biological process of genes, showing different expression levels between B. abortus infections. The different expression levels in B. abortus mutant strain infected RAW 264.7 cells were compared to wild-type infected cells at 24 h after infection. a Up-regulated genes. b Down regulated genes
The altered gene expressions in wild-type-infected RAW 264.7 cells compared to uninfected cells were also categorized by the PANTHER classification database. Following B. abortus wild-type infection, most of the genes showing different expression were up-regulated at 6 h post infection. Gene enrichment analysis revealed that these genes were associated with the immune system and cell defense processes (Ccl2, Gbp7, H2-T24, Ier3, Irg1, Irf7, Kdm6b, Ltb, Nfkbia, Nfkbiz, Oas2, Oasl2, Ptgs2, Slfn2, Tnf, Trim30a). Among these genes, Casp4, Ier3, Ifi204, Ltb, Nfkbia, Tnf, Xaf1, and Zc3h12a were also categorized to be associated with apoptosis. At 12 h post infection, RAW 264.7 cells showed down-regulation in 30 genes and up-regulation in 172 genes. The nucleosome core and DNA binding were found to be impacted by the down-regulated genes (Hist1h2ag, Hist1h2bj, Hist1h3g, Hist1h3c2, Serpinb1a, and Serpinb9b). Of the 172 genes up-regulated by Brucella infection, the 10 most up-regulated genes were associated with immune response (Irg1, Ifi44l, Ifit1, Ifi204, Oasl2, Ifi202b, H2-T24, Irf7, Cmpk2, and Usp18). In addition, genes associated with apoptosis were also up-regulated (Bcl2a1d, Cd40, Hck, Ltb, Nfkbia, Pim1, Rassf4, Stat1, Stat2, Tnf, Tnfrsf1b, Tnfrsf9). Maximum altered gene expression was observed after 24 h infection (Additional file 5: Figure S3 and Additional file 6: Figure S4). Genes showing altered expression were associated with 10 molecular functions and 14 biological processes. Most of the differentially expressed genes were involved in two molecular function categories (catalytic activity and binding) and two biological process categories (metabolic process and cellular process).
Analyses of affected pathways and gene networks following mutant strain infections
Pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database revealed that 24 h B. abortus infection led to cell cycle arrest. When compared to the wild type infected cells, no KEGG pathway was represented by more than 10 genes in the C24 and C30 mutant strain infected RAW 264.7 cells. Conversely, the cytokine-cytokine receptor interaction pathway was demonstrated by 11 down-regulated genes (Ccl2, Ccl5, Ccl7, Csf2, Csf3, Cxcl10, Cxcl11, Il1a, Il1b, Il6, and Tnfrsf1) in C3 mutant infected RAW 264.7 cells after 24 h. The TNF-like receptor pathway, chemokine-signal pathway, and Toll-like receptor pathway were found to be affected by altered genes in C3 mutant strain infected cells at 24 h. Among the differentially expressed genes in C3, C24, or C30 infected cells, one network was identified by the Ingenuity Pathway Analysis (IPA) in each mutant strain at 24 h after infection, as shown in Fig. 5. The network identified in C3 mutant strain infected cells was mainly associated with cytokine interactions among IL-6, IL-1A, and CCL2. In addition, gene networks of C24 and C30 mutant strains were on the prostaglandin-endoperoxide synthase 2 and tripartite-motif protein 30, respectively.
Identified network of the genes with altered expression in mutant strain infected RAW 264.7 cells. The different gene expression levels in B. abortus mutant strain infected RAW 264.7 cells at 24 h after infection were compared to the wild-type infected cells. Green color indicates down-regulation and arrows indicate directional relationships. a C3. b C24. c C30
Validation of microarray data
To verify the microarray results, gene expression levels of IL-1β, IL-6, Csf2, and Gadd45b in the microarray RNA samples were investigated by quantitative real time RT-PCR (qRT-PCR). The C3 mutant strain infected RAW 264.7 cells showed significant changes in gene expression level compared to wild-type infected cells. Validation was therefore performed based on these genes showing different expression levels. Furthermore, the microarray results revealed that pathways of “Responses of cytokine and immune defense”, and “Apoptosis process” were down-regulated in C3 mutant strain infected cells compared to wild-type infections. Hence, qRT-PCR for microarray validation was carried out using genes associated with these pathways (IL-1β, IL-6, Csf2, and Gadd45b). As shown Fig. 6, all genes evaluated by qRT-PCR showed similar expression levels as the microarray results.
Discussion
Brucella spp. are facultative intracellular bacteria like Salmonella enterica and Listeria monocytogenes. However, they express uncanonical virulence factors such as virulence regulator proteins and phosphatidylcholine, rather than classic virulence factors such as protease, exotoxin, cytolysins, and capules [1,2,3]. Moreover, Brucella spp. utilize and modulate normal functions of host cells, such as macrophages, to establish successful strategies for intracellular survival [5, 14, 15]. Therefore, characterization of the host macrophage-Brucella interaction is very important to identify the pathogenicity and infection mechanism of Brucella [22]. Microarray is a powerful approach that enables understanding the host responses at the global gene transcription level, thereby providing a plethora of information regarding the interactions associated with cell responses to antigens at the molecular level [15, 23]. In this study, microarray analysis was used to compare the responses between macrophages infected with B. abortus wild-type and B. abortus mutant strains, to demonstrate the Brucella genes that affect the interactions between host immune cells and the bacteria.
Previous studies of genes associated with pathogenicity of bacteria have been conducted using random insertion mutants generated by transposable elements [18, 19, 23, 24]. B. abortus mutant strains used in the current study were generated using transposon mutagenesis and single insertions certified in our previous study [18, 23, 25], in which Brucella genes associated with internalization and early host immune responses were investigated using 2 h infection experiments. In this study, we focused on the genes affecting interactions between host immune cells and Brucella, which would affect the survival strategy of the bacterium through longer infection period. Infection periods longer than 12 h are sufficient for Brucella to form replicative Brucella-containing vacuoles [26]. Among the mutant strains generated in our previous study [18], C3, C24, and C30 strains, which showed unique characteristics compared to the wild-type (Additional file 2: Figure S1 and Table 1), were selected for this study. The selection was carried out based on the growth rate and differences in responses of RAW 264.7 cells to infection with wild-type and mutant strains. The product levels of IL-6, TNF-α, and NO in infected cells, which are representative biomaterials associated with bactericidal responses of macrophage to bacterial infection [27,28,29], were used as criteria for identifying differences in responses of infected cells.
Macrophages have been reported to show early immune responses (such as inflammation) following B. abortus infection [6, 14, 15, 23, 30, 31]. Inflammation is a protective response to B. abortus infection observed in the early stages of infection, and coordinated by cytokines and chemokines [15, 30]. In this study, wild-type infected RAW 264.7 cells showed up-regulation in genes associated with immune responses (Ccl2, Gbp7, H2-T24, Ier3, Irg1, Irf7, Kdm6b, Ltb, Nfkbia, Nfkbiz, Oas2, Oasl2, Ptgs2, Slfn2, Tnf, Trim30a). Of these, Casp4, Ier3, Ifi204, Ltb, Nfkbia, Tnf, Xaf1, and Zc3h12a were also categorized as being associated with apoptosis, which plays a major role in responses to brucellosis by exposing the bacterium to the extracellular environments, where it encounters immune system components such as antibodies and the complement system, thereby reducing bacterial replication [6, 14, 15, 31]. In this study, most responses of RAW 264.7 cells at 6 h were focused on defense activities of macrophages. These results corresponded to the intracellular survival and replication after internalization in RAW 264.7 cells, demonstrated by a decrease in intracellular B. abortus wild-type at 6 h after internalization (Fig. 1b).
Following 12 h of infection, the nucleosome core and DNA binding were found to be down-regulated (Hist1h2ag, Hist1h2bj, Hist1h3g, Hist1h3c2, Serpinb1a, and Serpinb9b). Considering nucleosomes are selective products during apoptosis [32, 33], it is assumed that the apoptotic process of macrophages was affected by down-regulated genes due to Brucella infection. Interestingly, the most up-regulated genes were involved in the immune responses (Irg1, Ifi44l, Ifit1, Ifi204, Oasl2, Ifi202b, H2-T24, Irf7, Cmpk2, and Usp18), as were the genes associated with apoptosis (Bcl2a1d, Cd40, Hck, Ltb, Nfkbia, Pim1, Rassf4, Stat1, Stat2, Tnf, Tnfrsf1b, Tnfrsf9). These results indicate that the professional macrophages, namely the RAW 264.7 cells, elicited immune responses to clear Brucella infection consistently, concurrent with the activities of Brucella to elude the clearance efforts of the host [6, 14, 15, 23, 31, 34].
KEGG pathway mapping showed that B. abortus wild-type infected cells were down-regulated in cell cycle through down-regulation of pathways such as cell cycle (42 genes), purine metabolism (26 genes), pyrimidine metabolism (24 genes), and DNA replication (23 genes) at 24 h [35]. The p53 pathway, which inhibits cell proliferation and then induces apoptosis [36, 37], was also down-regulated. Those down-regulations were related with inhibition of cell death, which allows the survival of brucellae by avoiding exposure to the more hostile extracellular environment, thereby inducing more replication of B. abortus [31]. Moreover, it was observed that genes involved in phagosome formation were down-regulated (Scarb1, Tuba1a, Tuba4a, Tuba1c, Tubb5, Tubb4b, and Tubb6). Brucella is known to inhibit phagosome formation and phagosome-lysosome fusion to avoid host immune responses [9, 14, 38, 39]. In addition to these strategies, modification of cell cycle arrest, which may permit inhibition of apoptosis [15, 22], seemed to be more dominant than other down-regulated factors such as cytokine interaction and phagolysosome formation, considering the number of genes involved in cell arrest. Conversely, immune responses of RAW 264.7 cells to B. abortus infection for 24 h were focused on the cytokine interactions, as described in previous studies [11, 15, 31]. The up-regulation of the KEGG pathway of TNF-signaling and cytokine-cytokine receptor interaction was observed in this study. Moreover, these pathways involved genes that were differentially expressed by more than 20-fold, such as Ccf1, Il6, and Ccl5.
Although the overall transcriptional responses were similar between mutant strains and wild-type of B. abortus, notable difference were observed at the different time points. The 54 down-regulated genes in C3 mutants at 24 h were mainly involved in cell immune responses associated with cytokine-cytokine receptor interaction in the KEGG pathway database, which are mainly involved in immune responses elicited by RAW 264.7 cells to clear Brucella [4, 14, 15, 23]. Moreover, these genes were involved in the apoptosis process, which plays an important role in intracellular survival of Brucella [4, 15, 40]. The gene network analysis demonstrated that IL-1α, IL-6, tripartite-motif protein 30 (Trim30), prostaglandin-endoperoxide synthase 2 (PTGS2; also known as cyclooxygenase-2, COX-2), and CCL2 were down-regulated, showing interaction. These genes mediate the macrophage polarization to M1, a common response of macrophages to bacterial infections, which includes genes encoding TNF, IL-1β, IL-6, and CCL2 [41]. IL-6 plays a central role in the response of regulatory T cells to microbial infections, and is known to be a predominant mediator of the acute phase response in inflammation triggered by infection [42]. IL-6 synthesis is induced by COX-2 activation [42]. This positive relationship was found in the network of RAW 264.7 cells infected with C3 mutant strain. Several studies have proved the important role of IL-6 in Brucella infection. Indeed, IL-6 is known to play a role in the induction of acquired cellular resistance to intracellular bacteria by macrophage activation [43,44,45]. Important roles of IL-6 were also reported in the regulation of MHC II expression and antigen processing [45, 46]. Previous study further demonstrated that IL-6 inhibited intracellular replication through its control effects on endocytosis and endosome-phagosome fusion [47]. IL-1 has been reported to play an important role in protective responses against Brucella infection [48]. Trim30 regulates the NF-κB pathway as a negative feedback via the degradation of TAB2 and TAB3, resulting in inhibition of TRAF6 ubiquitylation, leading to the inhibition of NF-κB activity [49, 50]. Activation of the NF-κB pathway results in the induction of Type I IFN and other pro-inflammatory cytokines, thereby inducing protective effects against infection [51]. Down-regulation of Trim30d expression was presumed to be caused by lower NF-κB activity in C3 mutant strain infected cells. This down-regulation of genes associated with the protective immune responses against Brucella infection was confirmed by the quantification of NO, IL-6 and TNF-α in C3 mutant strain infected RAW 264.7 cells. In summary, the C3 mutant strain had the ability to inhibit the protective immune responses, via cytokine production.
The load of intracellular bacteria has differing effects on the gene expression in infected RAW 264.7 cells. However, there was no significant difference in the gene expression in the 12 h microarray analysis, between cells infected with wild-type and C24 mutant strains, although the RAW 264.7 cells were infected with higher CFU number of C24 mutants than wild-type strain (Fig. 1). In case of 24 h microarray analysis, even though differences in intracellular bacteria CFU number were observed among the strains of wild-type, C24, and C30 mutants, there was no significant difference of gene expression level in the infected RAW 264.7 cells compared to the wild-type infected cells. These results suggest that different gene expressions observed in the RAW 264.7 cells infected with C3 mutant compared to the cells infected with the wild-type were not significantly affected by the intracellular bacterial CFU load.
The ABC transporter permease (BruAb2_1031) has a role in dipeptide import [20]. Insertion of transposon into BruAb2_1031 was confirmed in the C3 mutant strain by BLASTIN analysis. The disruptive effects of BruAb2_1031 on the pathogenesis of B. abortus are not clearly understood. However, considering the important roles of peptide uptake in bacterial nutrition associated with growth and replication, especially in intracellular bacteria, it is plausible that mutation at BruAb2_1031 might affect the growth and virulence of B. abortus [52, 53]. These agreed with our results which showed slow growth and lower replication levels of C3 mutant in brucella broth and RAW 264.7 cells, respectively. Although the exact roles of the gene in B. abortus infection are not clear, this result implicated that mutation at BruAb2_1031 causes the inhibition in uptake of nutrition, thereby inducing the decrease of growth and intracellular replication.
Along with the effect on bacterial nutrition, disruption of ABC transporter affects the internalization and intracellular survival. Previous studies demonstrated that the mutation at genes associated with the transporter system (such as VirB, cgt, and cydC) reduced the internalization and intercellular survival in B. abortus [54,55,56]. These results implicate that disruption of the ABC transporter system generally exerts a negative effect on internalization, and the intracellular survival and replication of Brucella. However, a high level of intracellular survival was observed in the C3 mutant strain at 6 h after internalization, in spite of low levels of internalization and intracellular replication (Fig. 1) [6, 14, 15, 23, 30, 31]. This result was thought to be induced by reduced immune responses of RAW 264.7 cells to C3 mutant strain infection, which were agreeable with the results of microarray.
ABC permease utilizes the periplasmic binding protein (PBPs) to capture substrate and present it at the intake vestibules of the membrane translocator unit [57]. It is therefore presumed that the mutation at the BruAb2_1031 gene could induce changes in constituents of the bacterial envelope, even though they were small. This perturbation in bacterial envelope was speculated to alter the antigens interacting with macrophages, thereby reducing the immune responses of macrophage to infection of C3 mutant strain in this study [14, 31, 58]. This phenomenon was also supported by microarray analysis where down-regulation of the gene expression in C3 mutant infected RAW 264.7 cells was observed, especially in genes associated with M1 polarization of macrophages [58].
In the C24 mutant strain infected RAW 264.7 cells, most genes altered in expression were classified as predicted genes. The IPA network revealed the low immune response of RAW 264.7 cells to C24 mutant strain infection through down-regulation of COX-2, which is normally up-regulated in response to inflammatory and pro-inflammatory responses [59]. However, no other significant differences were observed in immune responses or apoptosis between RAW 264.7 cells infected with C24 mutant strain and wild-type strain. The C30 mutant infected RAW 264.7 cells also showed slight differences in gene expression compared to the wild-type infected cells. Similar to C24 mutant strain infected RAW 264.7 cells, most of the altered genes in the C30 mutant strain infected cells were predicted genes. There were no notable differences between cells infected with wild type or C30 mutant strain, except for down-regulation of Trim30. Therefore, further analyses on these predicted genes are required to demonstrate the role of BruAb2_0113 and ahpD in brucellosis.
Altogether, the C3 mutant strain showed high intracellular survival in RAW 264.7 cells compared to the wild-type. These cells also showed down-regulation of genes associated with protective immune responses to Brucella infection. Our results suggest that the C3 mutant strain has more enhanced strategies for intracellular survival than the wild-type. This enhanced intracellular survival ability could be presumed to be elicited by the mutation of BruAb2_1031, which is considered to be a cause of antigen alteration that could reduce the immune responses of macrophages in brucellosis.
The BruAb2_1031 gene (ABC transporter permease) is known to play a role in the transporter of peptides as a component of the ABC transporter [20]. However, our study showed BruAb2_1031 mutation is effective against Brucellosis. Our findings suggest that the ABC transporter permease could be a potential antigen in the development of Brucella vaccine in the future. In addition, to clarify a role of ABC transporter permease, changes in the function of ABC transporter by mutation of BruAb2_1031 gene need to be further investigated, and effects of BruAb2_1031 gene mutation on B. abortus should be identified.
Conclusions
Characteristics of mutant strains generated by transposon mutagenesis were investigated by infection experiments in RAW 264.7 cells. BLASTIN analysis revealed that the ABC transporter permease (BruAb2_1031) was mutated in the C3 mutant strain, which showed a higher intracellular survival rate in RAW 264.7 cells than the wild strain. This enhancement is presumed to be elicited by the mutation of BruAb2_1031, which is considered as a cause of antigen alteration that could reduce the immune responses of macrophages in brucellosis. Also, down-regulation of the protective immune response-related functions were observed in C3 mutant strain infected RAW 264.7 cells. This study reported for the first time that mutation of the BruAb2_1031 gene in B. abortus reduces the defense responses of RAW 264.7 cells to Brucella infection, indicating that the molecules associated with BruAb2_1031 gene have a role in immune responses of macrophages, and therefore its mutation enhanced the intracellular survival of B. abortus.
Methods
Bacterial strains and cell line
In our previous study, 132 mutant strains were generated from B. abortus 1119–3 strain (wild-type) [18, 23] using EZ-Tn5™ Transposome complexes (Epicentre® Biotechnologies, USA) [18]. Brucellae were cultured in brucella broth or agar (Difco, USA) and Brucella mutant strains were cultured with 30 μg/mL of kanamycin. The murine leukemic monocyte macrophage line, RAW 264.7, was obtained from the Korea Cell Line Bank (KCLB No.40071, Korea) and grown at 37 °C in 5% CO2 atmosphere in Roswell Park Memorial Institute medium (RPMI) 1640 (Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL). All procedures of the bacterial experiment were approved by the Seoul National University Institutional Biosafety Committee (SNUIBC-R160314–1).
Growth rates of B. abortus mutant strains
The bacterial growth rates were measured in brucella broth without antibiotics, according to the standard curve of CFU versus optical density. The level of bacterial growth rates was presented as the relative percentage compared to that of the wild-type, where the growth rate level of wild-type was regarded as 100%. Growth measurement experiments were independently conducted two times.
Selection of B. abortus mutant strains
Among the generated mutants by same electroporation of transposome, 26 mutant stains showed defective internalization (Additional file 2 Figure S1 and Additional file 1: Table S1). Because transposon mutagenesis could cause defective growth [60], the mutant strains were grouped based on the growth rate, as follows: group A of mutant strains showed more than 10% reduction in growth rate; group B of mutant strains showed similar growth rate compared to that of wild-type; group C of mutant strains showed more than 10% increment in growth rate. Among the mutant strains in each group, strains showing distinctive characteristics were selected for further studies.
The selection of mutant strains was carried out based on the differences in production levels of NO, IL-6, and TNF-α in RAW 264.7 cells responding to infection with each strain. Briefly, RAW 264.7 cells were cultured at a concentration of 8.0 × 104 cells/cm2 in 6-well culture plates, in 2 mL of media containing 2% FBS. After 8 h-culture, cells were infected with B. abortus wild-type or mutant strains at a multiplicity of infection (MOI) of 100 [18]. Following centrifugation at 150×g for 10 min at room temperature, the infected cells were incubated at 37 °C and 5% CO2 for 24 h. The NO production in supernatants of infected cells was measured using Griess reaction, as described previously [61]. The amounts of IL-6 and TNF-α in supernatants were measured using a cytokine ELISA kit (eBioscience, USA) according to the manufacturer’s instructions. All experiments were independently carried out twice.
Internalization, intracellular survival, and intracellular replication in RAW 264.7 cells
The levels of internalization, intracellular survival, and intracellular replication in RAW 264.7 cells were also investigated. RAW 264.7 cells were infected with B. abortus wild-type and mutant strains at an MOI of 100 [6, 18]. After 1 h, the medium was removed and replenished with medium containing 2% FBS and 30 μg/ml of gentamicin (gentamicin protection assay). The cells were then washed prior to lysis, and lysates were plated on to brucella agar at 0 h, 6 h, 12 h, 24 h, and 48 h after gentamicin treatment. The levels of internalization (0 h), intracellular survival (6 h) and replication for each strain (12 h, 24 h, and 48 h) were expressed by CFU. The CFU changes of intracellular bacteria were investigated through three repeated experiments.
Identification of transposon insertion sites
In our previous study, we had confirmed the single insertion of Tn5 transposome by Southern blot analysis [25]. The sequence of insertion site and disrupted gene in mutant strains were identified using PCR and alignment of search tool, as described previously [18]. DNA sequences from the PCR product were used to identify the insertion sites using analysis B. abortus chromosome II with circus (http://circos.ca/guide/genomic/).
Macrophage infection and RNA preparation for microarray
Macrophage infection was conducted using B. abortus wild-type and mutant strains at an MOI 10 [62, 63]. Following centrifugation at 150×g for 10 min at room temperature, the infected cells were incubated at 37 °C under 5% CO2 atmosphere for 6 h, 12 h, and 24 h. After three washes with DPBS (Gibco, USA), the RNA was extracted from the infected cells at each time point using an RNeasy mini kit (Qiagen, Netherlands), as described by the manufacturer. The purity and integrity of RNA was determined by denaturing gel electrophoresis, the OD260/280 ratio, and analysis on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). All RNA samples were quantified, divided into aliquots, and stored at − 80 °C until use.
Microarray hybridization
RNA amplification, labeling, array hybridization, and scanning were conducted by Macrogen Inc. (Seoul, Korea). The Affymetrix Whole Transcript Expression array process was conducted according to the manufacturer’s protocols using a GeneChip Whole Transcript PLUS reagent kit (Affymetrix, USA). Briefly, the GeneChip Whole Transcript Amplification kit (Affymetrix) was used for cDNA synthesis. The sense cDNA was then fragmented and biotin-labeled with terminal deoxynucleotidyl transferase, using a GeneChip Whole Transcript Terminal labeling kit (Affymetrix). Approximately 5.5 μg of labeled DNA target was hybridized to the Affymetrix GeneChip Mouse 2.0 ST Array for 16 h at 45 °C, which covers more than 35,000 transcripts. After washing step, hybridized arrays were stained on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner (Affymetrix). Signal values were computed using the Affymetrix® GenChip™ Command Console software (AGCC).
Raw data preparation and statistical analysis
Raw data extracted by AGCC were summarized and normalized by the robust multi-average (RMA) method implemented in Affymetrix® Expression Console™ Software. The results were exported with gene level RMA analysis, and subjected to the differentially expressed gene (DEG) analysis. Statistical significance of the expression data was determined using LPE test and fold change, in which the null hypothesis was that no difference exists among groups. False discovery rate was controlled by adjusting the p-value using the Benjamini-Hochberg algorithm. The genes that showed different expression levels in RAW 264.7 cells infected with B. abortus mutant strains compared to that of wild-type, were determined based on a p-value < 0.05 and a fold-change > |2|. Dataset of microarray results has been deposited in Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79264) and are accessible through GEO series accession number GSE79264.
Microarray data analysis
Genes showing significantly altered expressions were subjected to gene set enrichment analysis using the PANTHER classification database (http://www.pantherdb.org). Differently expressed genes were categorized by biological processes and molecular functions using the PANTHER classification database based on means of Fisher’s exact test, to identify coordinated changes in pre-defined sets of related genes.
The changes in cell process were derived from interactions among the differently expressed genes, which form the functional pathways and networks [64]. Therefore, genes showing altered expressions were analyzed using the KEGG database to identify systemic information representing functional aspects of each gene. For a pathway mapping term to be considered significant, pathways represented by fewer than 10 genes were filtered out for identification of the most affected pathways [65]. In addition, the Qiagen’s IPA (Ingenuity Systems Inc., USA) was performed for biological processes, canonical pathways, and networks analysis.
Verification of microarray results
To verify the microarray results, three genes associated with responses of cytokines and immune defense against Brucella infection, and one gene connected to the apoptosis process of the cell, were selected and subjected to qRT-PCR (Table 2). Total RNA was reverse transcribed using a QuantiTect Reverse Transcription kit (Qiagen), according to the manufacturer’s protocols. The qRT-PCR reaction was carried out with cDNA synthesized from 24 ng of RNA using the Rotor-Gene SYBR Green PCR kit (Qiagen) and Rotor-Gene Q real time PCR cycler (Qiagen), under the following conditions: 45 cycles at 95 °C for 15 s followed by 45 s at 60 °C [35]. The gene expression levels were analyzed by the 2-∆∆Ct method based on the house-keeping gene, β-actin, as a reference [35, 66].
Statistics
Data are presented as the mean ± standard deviation (SD). Statistical significance was analyzed by Student’s t-test or repeated measures of ANOVA (Tukey’s HSD test for post-test of multiple comparison) using SPSS version 23.0 software (SPSS, USA). The statistical significance of differences was set at value of p < 0.05.
Abbreviations
- CCL2:
-
Chemokine (C-C motif) ligand 2
- CFU:
-
Colony forming unit
- COX-2:
-
Cyclooxygenase-2
- DEG:
-
Differentially expressed gene
- DPBS:
-
Dulbecco’s Phosphate-Buffered Saline
- FBS:
-
Fetal bovine serum
- GRO-α:
-
Growth regulated oncogene alpha
- IFN-γ:
-
Interferon-gamma
- IL-12:
-
Interleukin-12
- IL-6:
-
Interleukin-6
- IL-8:
-
Interleukin-8
- IPA:
-
Ingenuity Pathway Analysis
- KCLB:
-
Korea Cell Line Bank
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LPS:
-
Lipopolysaccharide
- MCP-1:
-
Monocyte chemotactic protein 1
- MIP1 α/β:
-
Macrophage inflammatory protein alpha/beta
- MOI:
-
Multiplicity of infection
- NO:
-
Nitric oxide
- PANTHER:
-
Protein analysis through evolutionary relationships
- PCR:
-
Polymerase chain reaction
- PTGS2:
-
Prostaglandin-endoperoxide synthase 2
- qRT-PCR:
-
Quantitative real time polymerase chain reaction
- RANTES:
-
Regulated on activation, Normal T cell expressed and secreted
- RPMI:
-
Roswell Park Memorial Institute medium
- TNF-α:
-
Tumor necrosis factor alpha
- Trim30:
-
Tripartite-motif protein 30
References
Neta AVC, Mol JP, Xavier MN, Paixão TA, Lage AP, Santos RL. Pathogenesis of bovine brucellosis. Vet J. 2010;184:146–55.
Moreno E, Moriyón I. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc Natl Acad Sci. 2002;99:1–3.
Roop RM II, Gee JM, Robertson GT, Richardson JM, Ng WL, Winkler ME. Brucella stationary-phase gene expression and virulence. Annu Rev Microbiol. 2003;57:57–76.
Ficht TA. Intracellular survival of Brucella: defining the link with persistence. Vet Microbiol. 2003;92:213–23.
Skendros P, Pappas G, Boura P. Cell-mediated immunity in human brucellosis. Microbes Infect. 2011;13:134–42.
Gross A, Terraza A, Ouahrani-Bettache S, Liautard JP, Dornand J. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect Immun. 2000;68:342–51.
Porte F, Liautard JP, Köhler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun. 1999;67:4041–7.
Pizarro-Cerdá J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goñi I, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–24.
Byndloss MX, Tsolis RM. Brucella ssp. virulence factors and immunity. Annu Rev Anim Biosci. 2016;4:111–27.
Eskra L, Canavessi A, Carey M, Splitter G. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect Immun. 2001;69:7736–42.
Golding B, Scott DE, Scharf O, Huang LY, Zaitseva M, Lapham C, et al. Immunity and protection against Brucella abortus. Microbes Infect. 2001;3:43–8.
Zheng K, Chen DS, Wu YQ, Xu XJ, Zhang H, Chen CF, et al. MicroRNA expression profile in RAW264. 7 cells in response to Brucella melitensis infection. Int J Biol Sci. 2012;8:1013–22.
Jiang X, Leonard B, Benson R, Baldwin CL. Macrophage control of Brucella abortus: role of reactive oxygen intermediates and nitric oxide. Cell Immunol. 1993;151:309–19.
Pei J, Ficht TA. Brucella abortus rough mutants are cytopathic for macrophages in culture. Infect Immun. 2004;72:440–50.
Eskra L, Mathison A, Splitter G. Microarray analysis of mRNA levels from RAW264. 7 macrophages infected with Brucella abortus. Infect Immun. 2003;71:1125–33.
Sung KY, Jung M, Shin MK, Park HE, Lee JJ, Kim S, et al. Induction of immune responses by two recombinant proteins of Brucella abortus, outer membrane proteins 2b porin and cu/Zn superoxide dismutase, in mouse model. J Microbiol Biotechnol. 2014;24:854–61.
Ko J, Splitter GA. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin Microbiol Rev. 2003;16:65–78.
Cha SB, Rayamajhi N, Lee WJ, Shin MK, Jung MH, Shin SW, et al. Generation and envelope protein analysis of internalization defective Brucella abortus mutants in professional phagocytes, RAW 264.7. FEMS Immunol Med Microbiol. 2012;64:244–54.
van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11:435–42.
Jenner DC, Dassa E, Whatmore AM, Atkins HS. ATP-binding cassette systems of Brucella. Comp Funct Genomics. 2009;2009:354649.
Steele KH, Baumgartner JE, Valderas MW, Roop RM. Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J Bacteriol. 2010;192:4912–22.
He Y, Reichow S, Ramamoorthy S, Ding X, Lathigra R, Craig JC, et al. Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondrion-associated gene expression in mouse macrophages. Infect Immun. 2006;74:5035–46.
Cha SB, Lee WJ, Shin MK, Jung MH, Shin SW, Yoo AN, et al. Early transcriptional responses of internalization defective Brucella abortus mutants in professional phagocytes, RAW 264.7. BMC Genomics. 2013;14:426.
Vidal JE, Chen J, Li J, McClane BA. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One. 2009;4:e6232.
Park WB, Im YB, Jung M, Yoo HS. Molecular characteristics of Brucella abortus mutants generated using EZ-Tn5Tm pMODTm-3 transposon system. J Prev Vet Med. 2015;39:144–52.
Gorvel JP, Moreno E. Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol. 2002;90:281–97.
Wong SS, Zhou HR, Marin-Martinez M, Brooks K, Pestka JJ. Modulation of IL-1β, IL-6 and TNF-α secretion and mRNA expression by the trichothecene vomitoxin in the RAW 264.7 murine macrophage cell line. Food Chem Toxicol. 1998;36:409–19.
MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–50.
Hamza T, Li B. Differential responses of osteoblasts and macrophages upon Staphylococcus aureus infection. BMC Microbiol. 2014;14:207.
Chen F, Ding X, Ding Y, Xiang Z, Li X, Ghosh D, et al. Proinflammatory caspase-2-mediated macrophage cell death induced by a rough attenuated Brucella suis strain. Infect Immun. 2011;79:2460–9.
Roop RM II, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol. 2009;198:221–38.
Amoura Z, Chabre H, Koutouzov S, Lotton C, Cabrespines A, Bach JF, et al. Nucleosome-restricted antibodies are detected before anti-dsDNA and/or antihistone antibodies in serum of MRL-Mp lpr/lpr and +/+ mice, and are present in kidney eluates of lupus mice with proteinuria. Arthritis Rheum. 1994;37:1684–8.
Ren Y, Tang J, Mok M, Chan AW, Wu A, Lau C. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum. 2003;48:2888–97.
Rossetti C, Galindo C, Everts R, Lewin H, Garner H, Adams L. Comparative analysis of the early transcriptome of Brucella abortus–infected monocyte-derived macrophages from cattle naturally resistant or susceptible to brucellosis. Res Vet Sci. 2011;91:40–51.
Jung M, Shin MK, Jung YK, Yoo HS. Modulation of macrophage activities in proliferation, lysosome, and phagosome by the nonspecific immunostimulator, mica. PLoS One. 2015;10:e0117838.
Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908.
Hoe KK, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–36.
Harrison RE, Grinstein S. Phagocytosis and the microtubule cytoskeleton. Biochem Cell Biol. 2002;80:509–15.
Döhmer PH, Valguarnera E, Czibener C, Ugalde JE. Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell Microbiol. 2014;16:396–410.
Tian M, Qu J, Han X, Zhang M, Ding C, Ding J, et al. Microarray-based identification of differentially expressed genes in intracellular Brucella abortus within RAW264. 7 cells. PLoS ONE. 2013;8:e67014.
Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–9.
Chen BC, Liao CC, Hsu MJ, Liao YT, Lin CC, Sheu JR, et al. Peptidoglycan-induced IL-6 production in RAW 264.7 macrophages is mediated by cyclooxygenase-2, PGE2/PGE4 receptors, protein kinase a, IκB kinase, and NF-κB. J Immunol. 2006;177:681–93.
Zhan Y, Cheers C. Differential induction of macrophage-derived cytokines by live and dead intracellular bacteria in vitro. Infect Immun. 1995;63:720–3.
Delpino MV, Barrionuevo P, Macedo GC, Oliveira SC, Di Genaro S, Scian R, et al. Macrophage-elicited osteoclastogenesis in response to Brucella abortus infection requires TLR2/MyD88-dependent TNF-α production. J Leukoc Biol. 2012;91:285–98.
Xavier MN, Winter MG, Spees AM, Nguyen K, Atluri VL, Silva TM, et al. CD4+ T cell-derived IL-10 promotes Brucella abortus persistence via modulation of macrophage function. PLoS Pathog. 2013;9:e1003454.
Barrionuevo P, Cassataro J, Delpino MV, Zwerdling A, Pasquevich KA, Samartino CG, et al. Brucella abortus inhibits major histocompatibility complex class II expression and antigen processing through interleukin-6 secretion via toll-like receptor 2. Infect Immun. 2008;76:250–62.
Pizarro-Cerdá J, Desjardins M, Moreno E, Akira S, Gorvel JP. Modulation of endocytosis in nuclear factor IL-6 (−/−) macrophages is responsible for a high susceptibility to intracellular bacterial infection. J Immunol. 1999;162:3519–26.
Skyberg JA, Thornburg T, Kochetkova I, Layton W, Callis G, Rollins MF, et al. IFN-γ-deficient mice develop IL-1-dependent cutaneous and musculoskeletal inflammation during experimental brucellosis. J Leukoc Biol. 2012;92:375–87.
Bowie AG. TRIM-ing down tolls. Nat Immunol. 2008;9:348–50.
Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, et al. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–77.
McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23:46–56.
Braibant M, Gilot P, Content J. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol Rev. 2000;24:449–67.
Green RM, Seth A, Connell ND. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-Nitrosoglutathione. Infect Immun. 2000;68:429–36.
Roset MS, Ciocchini AE, Ugalde RA, de Iannino NI. Molecular cloning and characterization of cgt, the Brucella abortus cyclic β-1, 2-glucan transporter gene, and its role in virulence. Infect Immun. 2004;72:2263–71.
O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–20.
Truong QL, Cho Y, Barate AK, Kim S, Hahn TW. Characterization and protective property of Brucella abortus cydC and looP mutants. Clin Vac Immunol. 2014;21:1573–80.
Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682–99.
Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69.
Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188:21–8.
Kim S, Watarai M, Kondo Y, Erdenebaatar J, Makino SI, Shirahata T. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect Immun. 2003;71:3020–7.
Sreejayan, MNA R. Nitric oxide scavenging by curcuminoids. J Pharm Pharmacol. 1997;49:105–7.
Lee JJ, Kim DH, Kim DG, Lee HJ, Min W, Rhee MH, et al. Toll-like receptor 4-linked Janus kinase 2 signaling contributes to internalization of Brucella abortus by macrophages. Infect Immun. 2013;81:2448–58.
Pei J, Turse JE, Wu Q, Ficht TA. Brucella abortus rough mutants induce macrophage oncosis that requires bacterial protein synthesis and direct interaction with the macrophage. Infect Immun. 2006;74:2667–75.
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–60.
Waters KM, Masiello LM, Zangar RC, Tarasevich BJ, Karin NJ, Quesenberry RD, et al. Macrophage responses to silica nanoparticles are highly conserved across particle sizes. Toxicol Sci. 2009;107:553–69.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8.
Acknowledgements
The authors and this work was supported by NRF grant of MSIP (No. 2014R1A2A2A01007291), Korea Health Industry Development Institute (HI16C2130), BK21 PLUS and Research Institute for Veterinary Science, Seoul National University, Seoul, Republic of Korea.
Funding
This work was supported by NRF grant of MSIP (No. 2014R1A2A2A01007291), Korea Health Industry Development Institute (HI16C2130), BK21 PLUS and Research Institute for Veterinary Science, Seoul National University, Seoul, Republic of Korea. The funders had no role in study design, data collection and interpretation, writing the manuscript and the decision to submit the work for publication.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author HSY upon reasonable request.
Author information
Authors and Affiliations
Contributions
This study included the efforts of MJ involved in data analysis and drafting of manuscript. SS was involved in mutant strain characterization. YBI was involved in mutant strain characterization. WBP was involved in transposon insertion site identification. HSY was involved in experimental design and supervision of the experiments and manuscript revision. All authors read and approved the final manuscript and publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Growth of B. abortus wild-type and mutant strains at each time point. CFU was calculated using the standard curve of CFU versus optical density. (PDF 19 kb)
Additional file 2:
Figure S1. Characteristics of RAW 264.7 cells infected with B. abortus mutant strains. (a) Internalization was investigated using RAW 264.7 cells infected with B. abortus wild-type and mutant strains at an MOI of 100. (b), (c), (d) Product levels of NO, IL-6, and TNF-α in RAW 264.7 cells responding to infection with each strain (MOI 100) was measured 24 h after infection. Based on the growth rate, mutant strains in this study were divided into the three groups as follow: group A of mutant strains showed more than 10% reduction in growth rate; group B of mutant strains showed similar growth rate compared to that of wild-type; group C of mutant strains showed more than 10% increment in growth rate. In this study, RNA samples from the RAW 264.7 cells infected with mutant strain (C3, C24, and C30) were subjected to microarray analysis. The product level of IL-6 in RAW 264.7 cells infected with C3 and C24 mutant strains were close to or below detectable levels of the ELISA system. (TIF 939 kb)
Additional file 3:
Figure S2. The CFU numbers of intracellular B. abortus wild-type and mutant strains in RAW 264.7 cells. RAW 264.7 cells were infected with wild-type and each mutant strain for 1 h at MOI 100, after which a gentamicin protection assay was conducted. At the selected time points, the medium was removed and cells were washed prior to lysis; the lysate was then plated on to brucella agar. Intracellular CFU (Log10) numbers of each strain at selected time points after internalization was evaluated, which indicates the levels of intracellular survival (6 h) and replication (12 h, 24 h, and 48 h) at each time point after internalization in RAW 264.7 cells (*p < 0.05 and **p < 0.01). (TIF 503 kb)
Additional file 4:
Table S2. The genes showing altered expression in RAW 264.7 cells after B. abortus infection. The different expression levels in B. abortus infected RAW 264.7 cells were compared to uninfected cells. (PDF 1128 kb)
Additional file 5:
Figure S3. Categorization by molecular function of genes showing different expression levels after infection. The different expression levels in B. abortus wild-type and mutant strain infected RAW 264.7 cells were compared to uninfected cells. (a) Up-regulated genes. (b) Down regulated genes. (TIF 922 kb)
Additional file 6:
Figure S4. Categorization by biological process of genes showing different expression levels after infection. The different expression levels in B. abortus wild-type and mutant strain infected RAW 264.7 cells were compared to uninfected cells. (a) Up-regulated genes. (b) Down regulated genes. (TIF 936 kb)
Additional file 7:
Figure S5. Scatter plots showing different gene expressions. The different gene expression levels in B. abortus mutant strain infected RAW 264.7 cells were compared to cells infected with wild-type at 6 h, 12 h, and 24 h after infection. Genes showing different expression levels are indicated by red dots. (TIF 2217 kb)
Additional file 8:
Table S3. The genes showing altered expression in RAW 264.7 cells after C3 mutant strain infection. The different expression levels in B. abortus C3 mutant strain infected RAW 264.7 cells were compared to wild-type infected cells. (PDF 52 kb)
Additional file 9:
Table S4. The genes showing altered expression in RAW 264.7 cells after C24 mutant strain infection. The different expression levels in B. abortus C24 mutant strain infected RAW 264.7 cells were compared to wild-type infected cells. (PDF 41 kb)
Additional file 10:
Table S5. The genes showing altered expression in RAW 264.7 cells after C30 mutant strain infection. The different expression levels in B. abortus C30 mutant strain infected RAW 264.7 cells were compared to wild-type infected cells. (PDF 37 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/zero/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jung, M., Shim, S., Im, Y.B. et al. Global gene-expression profiles of intracellular survival of the BruAb2_1031 gene mutated Brucella abortus in professional phagocytes, RAW 264.7 cells. BMC Microbiol 18, 82 (2018). https://doi.org/10.1186/s12866-018-1223-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-018-1223-7