Abstract
Mangroves are unique intertidal ecosystems that provide ecological niches to different microbes, which play various roles in nutrient recycling and diverse environmental activities. The association between myxobacteria and mangroves are hitherto poorly understood. The aim of our study was to evaluate the myxobacterial community composition as well as isolate myxobacteria and to characterize the antimicrobial activity of myxobacteria isolates from Indonesian mangroves. Twenty-five cultivable myxobacteria were affiliated in six genera: Myxococcus, Corallococcus, Archangium, Chondromyces, Racemicystis and Nannocystis of the order Myxococcales based on partial 16S rRNA gene sequences. Thirteen crude extracts showed moderate activities against at least one of human pathogenic microorganisms. The crude extract of Racemicystis sp. strain 503MSO indicated a novel compound, which has not been reported in the database yet and the identification of this compound needs further study. The myxobacterial communities of three different sampling sites were analyzed using primers adapted for the myxobacteria group identification. The results showed that myxobacterial communities are more diverse than assumed. Therefore, our study has highlighted the importance of the mangrove habitat as promising harbor of myxobacteria as well as novel antimicrobial compounds with activity against pathogenic microorganisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mangroves are unique environments of small forest trees in brackish water with transitional zones in the coastal area. They tolerate a wide range of salinity, oxygen, nutrient and harbor many types of microorganisms [1, 2]. Different microorganisms origin from this habitat have been documented with different their bioactivity and compounds [3, 4]. However, there is a limited information about the antimicrobial potential of myxobacteria isolated especially from Indonesian mangroves, which are known as the world’s largest mangroves [5].
A preliminary study considered that marine environments are promising habitats for the isolation of antibiotic producing bacteria including myxobacteria. Myxobacteria are Gram-negative Deltaproteobacteria and it is one of the most fascinating groups producing natural microbial products [6, 7]. They produced more than a hundred new carbon skeletons and derivatives of compounds with antibacterial, antifungal, antimalarial, antioxidative, or antiviral activity [8,9,10,11]. Four genera of rare marine/saline origin are known so far and from two (Haliangium and Enhygromyxa) numerous (bioactive) secondary metabolites had been isolated like haliangicin, salimabromide, salimyxins, enhygrolides, and haliamide. Sharma & Thakur [13] noted that crude extracts with strong antimicrobial activities hint to the presence of promising antimicrobial compounds against infectious disease and treatment. Furthermore, these results were consistent with a previous study of Linares-Otoya et al. [12] on antimicrobial screening of predatory bacteria including myxobacteria from coastline. Therefore, it can be assumed that also saline environments like mangroves harbor an enormous potential of new antibiotic producing myxobacteria [14] and an exotic myxobacteria such as species Racemicystis for example from uncommon habitat should be explored.
Myxobacteria possess a social lifestyle with moving to get nutrients, preying to another microorganisms, and surviving from lack environment cooperatively in predatory groups [15]. Their communities are commonly found in various environments, even in extreme conditions [15,16,17]. A preliminary study considered the use of specific primers and probes to analyze myxobacterial communities in soil samples [18] in addition Li et al. [19] and Brinkhoff et al. [20] carried out PCR-DGGE (Denaturing Gradient Gel Electrophoresis) and quantitative real time PCR to define myxobacteria diversity. Clone bank analyses, cultivation approaches were also used to compare their diversity from different habitats [21]. Presently, next generation sequencing (NGS) of hypervariable regions of the 16S rRNA gene, is a tool that gives profound insights into microbial communities. Jiang et al. [22] stated tremendous success using this method in comparing the bacterial community diversity and composition between mudflat, edge, bulk, and rhizosphere of mangrove’s samples in Hong Kong, China. Furthermore, Linares-Otoya et al. [12] were able to detect predatory bacteria with NGS from the Peruvian coastline stating that the microbiome present in this region is a promising source for heterotrophic bacterial strains and has potential for bioprospecting of antibiotics. Nevertheless, there are no specific information about myxobacterial communities in mangrove habitat using the NGS method’s.
The use of culture-independent methods is of great interest, as it has identified a larger number of novel taxa when compared to culture-dependent methods under standard laboratory condition [12, 21,22,23]. However, the culture-dependent methods is important step to get novel antimicrobial compound, because without the successful isolated novel taxa from nature, the production of secondary metabolites never happened [14]. The aim of this study was to evaluate the myxobacterial community composition in Indonesian mangroves by Illumina sequencing of 16S rRNA genes using specific primers, to isolate myxobacteria, and to discover their potential for antimicrobial activity.
Material and Methods
Sample Collection
The mangrove samples were collected from Taman Muara Tawar, Bekasi (MT) (6°088772′N 107°002393′E), Taman Muara Angke, Jakarta (MA) (6°105321′N 106°735578′E), and Taman Mangrove Api-Api, Yogyakarta (MK) (7°894662′S 110°02554′E), at Java, Indonesia in November 2018 (Fig. S1). Total of six samples including sediment and leaf flakes were taken from each location. Approximately 50 g of the upper sediment with leaf, which fell on it, were placed in a sterile zip lock plastic bag. All samples were air dried at 30 °C for minimalizing contamination from fungi.
Isolation of Myxobacteria
The strains were isolated as previously described by Mohr et al. [21] using water agar with autoclaved Escherichia coli strain K12 and Stan 21 agar with filter paper [24]. Swarming colonies or fruiting bodies were observed under dissecting microscope (Olympus SZX10) every 5–15 days and transferred to new water agar plates with Escherichia coli strain K12 and finally to VY/2 agar plates [25]. The pure cultures from VY/2 agar plates were transferred into 20 ml CY/H liquid medium [24] and 1.5 ml of well-grown cultures were directly conserved at −80 °C.
Identification of Pure Cultures by 16S rRNA Gene Sequences
The bacterial DNA was extracted using Invitek Spin Plant Mini Kit (Invisorb) following the manufacturer’s instruction. One microliter of template DNA was directly amplified using F27/R1525 as described [21, 23]. The PCR products were checked by 0.8% agarose gel electrophoresis at 70 V for 40 min and purified by NucleoSpin Gel and PCR Clean up Kit (Macherey–Nagel, Düren, Germany). The forward and reverse sequences of 16S rRNA gene fragments were assembled with the BioEdit program [26] and closely related type strains were identified using the NCBI 16S rRNA gene database. Sequences were deposited at NCBI database under accession number MW199130, MW182265-MW182288.
Microbiome Analysis
The total DNA was extracted from 250 mg of sample using the MOBio PowerSoil® Kit following the manufacturer’s instructions. The quality of DNA was measured in Nano-photometer IMPLAN. Amplification was performed using PrimerSTAR HS DNA Polymerase (Takara, Otsu, Shigu, Japan) following the manufacturer’s instructions. Amplification for analyzing the microbial community composition was described by Rath et al. [27] with modifications. Forward primers W2 and W5 [18] specifically targeting Cystobacterineae and Sorangiineae/Nannocystineae (Table S1) of the Myxococcales were separately used in conjunction with reverse primer R1525 [21, 23] in a first PCR reaction. Samples were pre-denaturated at 96 °C for 3 min following 25 cycles of denaturation at 94 °C for 1 min, annealing at 56.6 °C for 1 min, extension at 72 °C for 2 min. The PCR product was checked on 2.0% agarose gel electrophoresis and purified using NucleoSpin Gel PCR Clean up Kit (Macherey–Nagel, Düren, Germany). One microliter of purified PCR product was used as template in a second PCR with primers 807F and 1050R containing part of the sequencing primer sites as short overhangs (Table S1) for 20 cycles to enrich for target sequences. A third amplification step of 10 cycles added the two indices and Illumina adapters to amplicons [27]. The second and third PCR reaction following a first step PCR reaction. Obtained products were pooled in equimolar ratios and sequenced on an Illumina Miseq (2 × 300 bases, San Diego, USA).
The bioinformatic processing was performed as previously described [28]. Raw reads were merged with the Ribosomal Database Project (RDP) assembler [29]. Sequences were aligned within MOTHUR [30] (gotoh algorithm using the SILVA database [31] and subjected to preclustering (diffs = 2) yielding so-called phylotypes that were filtered for an average abundance of ≥ 0.001% and a sequence length ≥ 250 bp before analysis. The potential duplications of same Myxococcales sequences between both sets of primers pair were counted and were included to each primer pair analysis. Phylotypes were assigned to a taxonomic affiliation based on the naive Bayesian classification with a pseudo-bootstrap threshold of 80% [29, 32]. All sequences not matching to the order Myxococcales were deleted before further analysis. The relative abundance of genera was plotted using pivot table on Microsoft Excel. Sequences were deposited at NCBI database under accession number SRX9502207-SRX9502210 (Project PRJNA678217).
Preparation for Crude Extracts
Seed cultures from the 25 isolates were prepared from myxobacterial swarming colonies after seven and before 15 days of incubation (depending on the myxobacteria growth rate) on VY/2 agar medium [24] by inoculation into 20 ml CY/H liquid medium [24]. The cultures were shaken at 160–180 rpm. After seven days, 10% of the seed cultures were transferred into 100 ml screening VY/2 liquid medium [24], which contained 2% XAD-absorber resin and incubated at room temperature for 7 up to 15 days. After 2% XAD-absorber was collected, eluate molecules bound on XAD was released using 70 ml using 70 ml acetone (J. T. Baker) and the organic phase was evaporated (Heidolph, Laborota 4003 control) at a temperature of 38–40 °C. The residue was mixed with 1 ml methanol (J. T. baker) and stored in a freezer at −20 °C.
Screening for Antimicrobial Activity
All 25 crude extracts from 25 strains were evaluated by a test panel using the following pathogenic microorganisms: Escherichia coli WT-BW 25113, Escherichia coli JW0451-02, Acinetobacteria baumanii DSM 30008, Citrobacter freundii DSM 30039, Pseudomonas aeruginosa Pa14, Staphylococcus aureus Newmann, Mycobacterium smegmatis ATCC 700084, Candida albicans DSM 1665, Wickerhamomyces anomalus DSM 6766, and Mucor hiemalis DSM 2656. The antimicrobial activity was assessed by a growth inhibition test using a serial dilution of each crude extract against different pathogenic microorganisms in a 96-well plate [33]. The highest dilution, at which growth inhibition was observed, was noted. The antimicrobial activity was visualized quantitatively by heat map with Heatmapper software [34].
Identification of Active Compound
The crude extract of Racemicystis sp. strain 503MSO was fractionated on 96-well plate to analyze the peak-activity correlation. Separation was conducted on an Agilent 1260 high performance liquid chromatography (HPLC) (Agilent Technology USA) using XBridge C18 column (Waters, Milford, Massachusetts, USA) (100 × 2.1 mm). The solvents consist of 5% acetonitrile (MeCN) in water (H2O) added with 5 mmol ammonium acetate (NH4OAc) and 0.04 ml/l acetic acid (CH3COOH) as solvent A and solvent B consist of 95% MeCN, 5 mmol (NH4OAc,) and 0.04 ml/l CH3COOH. Gradient system starting from 10% B to 100% B in 30 min and maintaining at 100% for 10 min, followed by post-run in 10 min for column re-conditioning. Flow rate was 0.3 ml/min and column temperature maintained at 40 °C. The fractions was collected every 30 s [35]. In paralel, the crude extract was analyzed by using diode array detector—high-resolution electrospray ionization mass-spectrometry (DAD-UV-HRESIMS) (MaXis ESI TOF, Bruker Daltonik GmbH, Bremen, Germany) using bridge ethylene hybrid (BEH) C18 column (Waters ACQUITY, Milford, MA, USA) (1.7 µm 2.1 × 50 mm). A linear gradient from 95% H2O and 5% MeCN to 5%H2O and 95% MeCN suplemented with 0.1% formic acid was used [33]. The eluate on 96-well plate was dried under streaming nitrogen (N2) gas and the respective bacteria was incubated directly to the plate. The clear zone on the well plate indicating the active compounds as well as the retention time in the respective mass chromatogram. The data were analyzed following Krug and Müller [36] and Hoffmann et al. [37] using Data Analysis 4.2 B383 (Bruker Daltonics) and identified with the in-house compounds library (Myxobase).
Statistical Analysis
The results were analyzed using The IBM SPSS Statistics version 26.0 [38]. It was expressed as mean ± SEM (Standard Error of Mean). The differences between mean were statistically analyzed using one-way analysis of ANOVA with test of homogeneity of variances Bonferroni. P < 0.05 considered to statistical significance.
Results
Mangrove Myxobacteria Isolates
Seventy strains were successfully isolated from three different sources of Indonesian mangroves. All of the strains showed fruiting bodies formation and swarming on a surface agar medium. Therefore, based on different morphologies and 16S rRNA gene sequences analysis, the number of myxobacteria replicates from same source of organisms and location were reduced and 25 of 70 isolates were obtained. Twenty-five isolates, 16 from MA sampling site, 3 from MT sampling site and 6 from MK sampling site were selected for further analysis. Based on 16S rRNA gene sequence analysis, members of three suborders of Myxococcales were identified (see Table 1). Some isolates showed less than 98.60% similarity to close related type strains. Therefore, full genome sequencing is needed for their further characterization and whether they comprise novel species.
As main characteristics of myxobacteria as previously described by Reichenbach [16, 25], some of the swarming and fruiting bodies of myxobacterial isolates from mangroves can be seen in Fig. 1. Myxococcus sp. strains 431MSO and 451MSO have spherical fruiting bodies with yellow or oranges red colors on VY/2 agar medium. Coralococcus sp. strain 412MSO move with swarm colonies and forms fruiting bodies with coralloid-branched shapes. Chondromyces sp. strain 151MSO builds tree shaped fruiting bodies on Stan21 [39] agar medium and Archangium sp. strain 455MSO makes swarm colonies with branched radial veins on VY/2 agar medium. Racemycistis sp. strain 503MSO has swarming area like the genus Sorangium on VY/2 agar medium.
Myxobacterial Diversity
Myxobacterial diversity was evaluated after amplification using two different primer pairs W2/R1525 and W5/R1525, targeting the suborders Cystobacterineae and Sorangiineae/Nannocystineae, respectively [18, 21, 23]. A total of 20,057 myxobacterial sequences (1761 ± 1002 Standard Error of Mean/SEM per sample) from 12 samples, where each sample was amplified twice with the two different primer pairs, were obtained. The success of the W2/R1525 primer to amplify members of the order Myxococcales order varied and between 0.56% and 70.42% of the obtained reads belonged to this order (see Table 2). With the W5/R1525 primer pair, 7.27–27.03% of amplified sequences were belonging to Myxococcales, indicating a low abundance of those bacteria in that sample. In contrast to the W5/R1525 primer pair, 70.42% of Myxococcales sequences with the W2/R1525 primer pair in one Yogyakarta mangrove sample indicate high abundance of Myxococcales. Moreover, sample MK from Yogyakarta contained more diverse myxobacteria than other and sample MT from Bekasi showed less diversity according to the results.
Thirteen major genera were identified from three sampling sites analyzed in this study (Fig. 2). The W2/R1525 primer pair could also amplify sequences indicating the presence of Cystobacter, Myxococcus, Stigmatella, Archangium and Anaeromyxobacter of the Cystobacterineae. However, also Haliangium sequences was amplified by this primer. The W5/R1525 primer pair amplified sequence indicating the presence of members of the genera Haliangium, Kloferia and Nannocystis of the Nannocystineae and Chondromyces, Labilithrix, Phaselicystis, Polyangium and Sandaracinus of the Sorangiineae. In addition, some sequences of Myxococcus, Stigmatella and Cystobacter of the Cystobacterineae were observed as being amplified by this primer.
Relative abundances of myxobacterial genera in Indonesian Mangroves. MT: Bekasi MA: Jakarta MK: Yogyakarta using different set of primer W2/R1525 and W5/R1525. The community composition was revealed using primers targeting the suborder Cystobacterineae and the suborders Sorangiineae/Nannocystineae, respectively
Overall, the primers showed good specificity for their targets. Clear differences in myxobacterial diversity could be observed in the sampling sites. Stigmatella spp. exclusively existed in MK samples, Yogyakarta mangroves whereas Nannocystis spp., and Labilithrix spp. were present only in Jakarta and Yogyakarta mangroves. Only Myxococcus spp. were observed in all six samples, indicating that it is a common myxobacterium in mangroves samples (Table 3).
Antimicrobial Activity of Myxobacteria
Twenty-five crude extracts from the 25 isolates were tested for their antimicrobial activity against ten pathogenic microorganisms, including five isolates Gram-negative bacteria, two-isolates Gram-positive bacteria and three fungi isolates (Table 3). Thirteen crude extract showed activity against at least one of the human pathogenic microorganisms tested.
Two of them showed activity against both Gram-negative strains Escherichia coli and Citrobacter freundii and nine were active against Gram-positive Staphylococcus aureus. Furthermore, three of the extracts showed activity against the yeasts Candida albicans and Wickerhamomyces anomalus.
The crude extract of Racemicystis sp. strain 503MSO was fractionated in order to identify the responsible active compounds. We considered and selected Racemicystis sp. strain 503MSO out of 13 active strains because of ≤ 98.60% 16S rRNA sequence similarity to the type strain and because of missing information about compounds from genus Racemicystis in the Myxobase database.
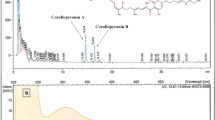
The crude extract of Racemicystis sp. strain 503MSO was fractionated in order to identify the responsible active compounds. The compound identification was done by comparing detected mass of the parent ions of the active fractions with in-house Myxobase database. The Myxobase is a database to support research with myxobacteria, which are increasingly recognized as producers of secondary metabolites. Within Myxobase, the information of bioactivity, retention time, UV spectrum, molecular mass and elemental formula of the molecule responsible for the active peak and HPLC chromatogram were provided. A summary of the active compounds from strain 503MSO was provided in Figure S1.
Two active fractions from Racemicystis sp. strain 503MSO were identified comprising compounds with masses of m/z 375.2531 and 604.3857 [M + H]+, respectively after the high-resolution mass-spectrometry (HR-MS) analysis of these active fractions. The peak chromatogram which indicated the presence of compound from the crude extract was mentioned in Fig. S1.
Discussion
Due to the increasing need for new antibiotics, the present study focused on the evaluation of the myxobacterial community as well as the isolation of novel members of the order Myxococcales and their potential for antimicrobial activity. Recent isolations of members of the myxobacteria have frequently resulted in the identification of new secondary metabolites with antibiotic activities. Untapped habitats such as various marine ecosystem have been proven to be a valuable resources for new microorganisms [7, 40, 41]. Indonesian mangroves explored in the present study were unexploited marine habitats and have a huge potential for the isolation of new uncommon species of myxobacteria.
Twenty-five strains were isolated from three different sites of Indonesian mangroves by conventional methods for isolation. Myxococcus strains were presented in all six samples and it reflected both myxobacteria isolates versus myxobacterial communities that were amplified based on 16S rRNA gene sequences. According to the fruiting body formation of Myxococcus, they were easy to be purified and cultivated under laboratory conditions [42]. Previously, this genus was used as model for wide-ranging studies of biotechnology [15, 43,44,45,46].
The analysis of myxobacterial communities by culture-independent methods revealed more abundances than myxobacterial isolates by culture-dependent method. For instance, the culture-independent community analyses revealed the genus Haliangium to be one of the most dominant and common myxobacteria in MA and MT samples, whereas it was not found by the culture-dependent method in our study. The faster growing fungal or bacterial contaminants exhaust nutrients before slow growing myxobacteria could be observed and further, the insufficient growth conditions in laboratory missing requirements such as changing tidal-caused salinity might be reasons that rare myxobacterial species were not isolated [21].
Thirteen out of 25 crude extracts from myxobacterial isolates yielded different activities against pathogenic microorganisms. These predatory bacteria including myxobacteria from coastline may be promising sources for novel antibiotics [12].
In this study, we only focused on the active crude extract of Racemicystis sp. strain 503MSO, an isolate from Yogyakarta mangroves. Racemicystis sp. strain 503MSO was selected for further analysis because there was no information about potential bioactive compounds from the genus Racemicystis in the Myxobase database yet. Therefore, it has a higher possibility to get new compounds from this genus.
The active fraction of Racemicystis sp. strain 503MSO with m/z 375.2531 [M + H]+ and 604.3847 [M + H]+ were detected in this study and may represent so far, an undescribed compound. Therefore, Racemicystis sp. strain 503MSO should be further explored including substantial work for optimizing isolation, structure elucidation and biological activity of the pure substance. To the best of our knowledge no bioactive compounds from the same genus and other sources were reported in the scientific publication to date.
Indonesian mangroves were inhabited by complex myxobacterial communities as shown by amplification using primer targeting Cystobacterinae (W2/R1525) and Sorangiineae/Nannocystineae (W5/R1525) [18, 21, 23]. Previously, Linares-Otoya [12] observed that the use of universal primers (F515/R806) yielded only a neglectable amount of myxobacterial sequences (equivalent to 0.0002%) as also indicated by our work.
The rapid sequencing-based analysis of myxobacterial communities has advantages compared to previous studies comprising DGGE (Denaturing Gel Gradient Electrophoresis) [47], hybridization analysis of a 16S rRNA gene library [18] or library analysis [21, 23] and by use of two PCR reactions an overview of all three suborders, i.e., Cystobacterineae, Sorangiineae and Nannocystineae were obtained [48, 49]. However, with the current method, a high amount of non-Myxococcales genera were amplified and target sequences often comprised < 10% of sequences. Evidently, optimization of primers is still necessary. Moreover, a species level information is only possible in exceptional cases using the V5V6 hypervariable region as performed here.
Overall, mangrove habitats are a rich source for antimicrobial compounds produced by myxobacteria, highlighting the necessity to explore their diversity.
Conclusions
This study confirmed that neglected areas such as mangroves are promising habitats for the isolation of novel myxobacteria strains and potential sources for unknown secondary metabolites producers. Based on comparison of its monoisotopic mass and retention time of compound described in Myxobase database, we suggest further studies for isolating and upscaling active compound of Racemicystis sp. strain 503MSO and applying novel models of biological test as well.
Data Availability
The data presented in this study are available in the Supplementary Information and at NCBI database under accession number SRX9502207-SRX9502210 (Project PRJNA678217).
Code Availability
The authors declared that the consent for code availability is not applicable.
References
Jiang Z-K, Tuo L, Huang D-L et al (2018) Diversity, novelty, and antimicrobial activity of endophytic actinobacteria from mangrove plants in Beilun Estuary National Nature Reserve of Guangxi, China. Front Microbiol 9:1–11. https://doi.org/10.3389/fmicb.2018.00868
Li F, Liu S, Lu Q et al (2019) Studies on antibacterial activity and diversity of cultivable actinobacteria isolated from mangrove soil in futian and maoweihai of China. Evidence-based Complement Altern Med. https://doi.org/10.1155/2019/3476567
Isaka M, Suyarnsestakorn C, Tanticharoen M et al (2002) Aigialomycins A-E, new resorcylic macrolides from the marine mangrove fungus Aigialus parvus. J Org Chem 67:1561–1566. https://doi.org/10.1021/jo010930g
Retnowati W (2010) Identification of Streptomyces sp-MWS1 producing antibacterial compounds. Indones J Trop Infect Dis 1:82. https://doi.org/10.20473/ijtid.v1i2.2171
Murdiyarso D, Purbopuspito J, Kauffman JB et al (2015) The potential of Indonesian mangrove forests for global climate change mitigation. Nat Clim Chang 5:1089–1092. https://doi.org/10.1038/nclimate2734
Schäberle T-F, Goralski E, Neu E et al (2010) Marine myxobacteria as a source of antibiotics—comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Mar Drugs 8:2466–2479. https://doi.org/10.3390/md8092466
Schäberle T-F, Lohr F, Schmitz A, König GM (2014) Antibiotics from myxobacteria. Nat Prod Rep 31:953–972. https://doi.org/10.1039/c4np00011k
Gerth K, Pradella S, Perlova O et al (2003) Myxobacteria: Proficient producers of novel natural products with various biological activities—Past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol 106:233–253
Wenzel S-C, Müller R (2009) Myxobacteria—“microbial factories” for the production of bioactive secondary metabolites. Mol Biosyst 5:567–574. https://doi.org/10.1039/b901287g
Landwehr W, Wolf C, Wink J (2016) Actinobacteria and myxobacteria–-two of the most important bacterial resources for novel antibiotics. In: Stadler M, Dersch P (eds) How to overcome the antibiotic crisis: facts, challenges, technologies and future perspectives. Springer International Publishing, Cham, pp 273–302
Mulwa L, Stadler M (2018) Antiviral Compounds from myxobacteria. Microorganisms 6:73. https://doi.org/10.3390/microorganisms6030073
Linares-Otoya L, Linares-Otoya V, Armas-Mantilla L et al (2017) Diversity and antimicrobial potential of predatory bacteria from the peruvian coastline. Mar Drugs 15:1–14. https://doi.org/10.3390/md15100308
Sharma P, Thakur D (2020) Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci Rep 10:1–18. https://doi.org/10.1038/s41598-020-60968-6
Mohr K (2018) Diversity of myxobacteria—we only see the tip of the iceberg. Microorganisms 6:84. https://doi.org/10.3390/microorganisms6030084
Muñoz-Dorado J, Marcos-Torres F-J, García-Bravo E et al (2016) Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol. https://doi.org/10.3389/fmicb.2016.00781
Reichenbach H (1999) The ecology of the myxobacteria. Environ Microbiol 1:15–21. https://doi.org/10.1046/j.1462-2920.1999.00016.x
Dawid W (2000) Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev 24:403–427
Wu Z-H, Jiang D-M, Li P, Li Y-Z (2005) Exploring the diversity of myxobacteria in a soil niche by myxobacteria-specific primers and probes. Environ Microbiol 7:1602–1610. https://doi.org/10.1111/j.1462-2920.2005.00852.x
Li S-G, Zhou X-W, Li P-F et al (2012) The existence and diversity of myxobacteria in lake mud—a previously unexplored myxobacteria habitat. Environ Microbiol Rep 4:587–595. https://doi.org/10.1111/j.1758-2229.2012.00373.x
Brinkhoff T, Fischer D, Vollmers J et al (2012) Biogeography and phylogenetic diversity of a cluster of exclusively marine myxobacteria. ISME J 6:1260–1272. https://doi.org/10.1038/ismej.2011.190
Mohr K-I, Zindler T, Wink J et al (2017) Myxobacteria in high moor and fen: an astonishing diversity in a neglected extreme habitat. Microbiologyopen 6:1–9. https://doi.org/10.1002/mbo3.464
Jiang X-T, Peng X, Deng G-H et al (2013) Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol 66:96–104. https://doi.org/10.1007/s00248-013-0238-8
Mohr K-I, Stechling M, Wink J et al (2016) Comparison of myxobacterial diversity and evaluation of isolation success in two niches: Kiritimati Island and German compost. Microbiologyopen 5:268–278. https://doi.org/10.1002/mbo3.325
Dworkin M (2007) Myxobacteria. In: eLS. https://doi.org/10.1002/9780470015902.a0020391
Reichenbach H (2006) The order cytophagales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes. Springer, New York, NY. https://doi.org/10.1007/0-387-30747-8_2
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Rath S, Heidrich B, Pieper D-H, Vital M (2017) Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 5:1–14. https://doi.org/10.1186/S40168-017-0271-9
Schulz C, Schütte K, Koch N et al (2018) The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 67:216–225. https://doi.org/10.1136/gutjnl-2016-312904
Wang Q, Garrity G-M, Tiedje J-M, Cole J-R (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Schloss P-D, Westcott S-L, Ryabin T et al (2009) Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
Webb G-I (2010) Naïve Bayes. In: Sammut C, Webb GI (eds) Encyclopedia of machine learning. Springer, Boston, pp 713–714
Wibowo J-T, Kellermann M-Y, Versluis D et al (2019) Biotechnological potential of bacteria isolated from the sea cucumber Holothuria leucospilota and Stichopus vastus from Lampung, Indonesia. Mar Drugs 17:635
Babicki S, Arndt D, Marcu A et al (2016) Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44:W147–W153. https://doi.org/10.1093/nar/gkw419
Primahana G, Risdian C, Mozef T et al (2020) Nonocarbolines A-E, β-carboline antibiotics produced by the rare actinobacterium nonomuraea sp. From Indonesia Antibiotics. https://doi.org/10.3390/antibiotics9030126
Krug D, Müller R (2014) Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep 31:768–783. https://doi.org/10.1039/c3np70127a
Hoffmann T, Krug D, Bozkurt N et al (2018) Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat Commun 9:1–10. https://doi.org/10.1038/s41467-018-03184-1
George D (2020) IBM SPSS statistics 26 step by step: a simple guide and reference/Darren George, Paul Mallery, 16th edn. Routledge, NY
Shimkets L-J, Dworkin M, Reichenbach H (2006) The Myxobacteria. In: Dworkin M, Falkow S, Rosenberg E et al (eds) The prokaryotes : volume 7: proteobacteria: delta, epsilon subclass. Springer, NY, pp 31–115
Albataineh H, Cole Stevens D (2018) Marine myxobacteria: a few good halophiles. Mar Drugs. https://doi.org/10.3390/md16060209
Sun Y, Feng Z, Tomura T et al (2016) Heterologous production of the marine myxobacterial antibiotic haliangicin and its unnatural analogues generated by engineering of the biochemical pathway. Sci Rep 6:1–11. https://doi.org/10.1038/srep22091
Zhang X, Yao Q, Cai Z et al (2013) Isolation and identification of myxobacteria from saline-alkaline soils in Xinjiang. China PLoS One 8:e70466
Keane R, Berleman J (2016) The predatory life cycle of Myxococcus xanthus. Microbiol (UK) 162:1–11. https://doi.org/10.1099/mic.0.000208
Wallace R-A, Black W-P, Yang X, Yang Z (2014) A CRISPR with roles in Myxococcus xanthus development and exopolysaccharide production. J Bacteriol 196:4036–4043. https://doi.org/10.1128/JB.02035-14
Korp J, Winand L, Sester A, Netta M (2018) Engineering pseudochelin production in Myxococcus xanthus. Appl Environ Microbiol 84:1–12. https://doi.org/10.1128/AEM.01789-18
Sudo S, Dworkin M (1972) Bacteriolytic enzymes produced by Myxococcus xanthus. J Bacteriol 110:236–245. https://doi.org/10.1128/jb.110.1.236-245.1972
Li B, Yao Q, Zhu H (2014) Approach to analyze the diversity of myxobacteria in soil by semi-nested PCR-denaturing gradient gel electrophoresis (DGGE) based on taxon-specific gene. PLoS ONE 9:1–8. https://doi.org/10.1371/journal.pone.0108877
Fanning S, Proos S, Jordan K, Srikumar S (2017) A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front Microbiol 8:1–16. https://doi.org/10.3389/fmicb.2017.01829
Black J-S, Salto-Tellez M, Mills K-I, Catherwood M-A (2015) The impact of next generation sequencing technologies on haematological research—a review. Pathogenesis 2:9–16. https://doi.org/10.1016/j.pathog.2015.05.004
Acknowledgements
We are grateful thank to Dr. Kathrin I Mohr for fruitful discussion and to Iris Plumeier, Silke Kahl, Klaus Peter Conrad, Birte Trunkwalter, and Stephanie Schulz for providing technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. This research was supported and fully funded by the German Academic Exchange Service (DAAD)-GINAICO program number 57342738 (91654412) for the PhD scholarship.
Author information
Authors and Affiliations
Contributions
Conceptualized: SO, JW, DP. Supervised: JW, DP. Performed the experiments: SO. Analyzed the data: SO, GP, LB. Wrote the paper-original draft preparation: SO. Review and edited: SO, GP, LB, DP, TM, and JW. Senlie Octaviana and Gian Primahana are equally contributed.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of this study; and in the collection, analyses, data interpretation, manuscript writing, or in the decision to publish the results.
Ethical Approval
This research does not involve human participants and/or animals. Therefore, no ethical approval is involved.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Octaviana, S., Primahana, G., Mozef, T. et al. Diversity of Myxobacteria Isolated from Indonesian Mangroves and Their Potential for New Antimicrobial Sources. Curr Microbiol 80, 46 (2023). https://doi.org/10.1007/s00284-022-03066-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03066-2