Abstract
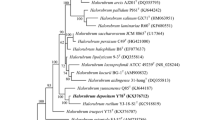
A halophilic archaeon, designated strain LS1_42T, was isolated from Sambhar Salt Lake, Rajasthan, India. Cells were non-motile, coccoid, Gram-stain-variable and present in irregular clusters with light pink pigmented colonies. The strain was strictly aerobic and able to grow without Mg2+. Growth of the strain LS1_42T was observed at 25–45 °C, pH 7.0–11.0 and NaCl concentrations of 10–35% (w/v). The nearest phylogenetic neighbor of strain LS1_42T was Natronococcus amylolyticus Ah-36T based on 16S rRNA and rpoB′ genes with similarity of 95.4% and 91.9%, respectively. Phylogenetic analysis based on 16S rRNA gene, rpoB′ gene and whole-genome sequences indicate that the strain LS1_42T belongs to the genus Natronococcus and is closely related to N. amylolyticus. The genome size was 5.38 Mb with 98.9% completeness. The DNA G + C content of the strain LS1_42T was 63.0 mol%. The average nucleotide identity, average amino acid identity and DNA–DNA hybridization values between LS1_42T and N. amylolyticus Ah-36T were 81.3%, 77.7% and 24.8%, respectively. The major polar lipids detected were phosphatidylglycerol and phosphatidylglycerol phosphate methyl ester. On the basis of phenotypic, chemotaxonomic and genome-based analysis, strain LS1_42T represents a novel species within the genus Natronococcus, for which the name Natronococcus pandeyae sp. nov. is proposed. The type strain is LS1_42T (MCC 3654T = JCM 33003T = KCTC 4280T = CGMCC 1.16738T).



Similar content being viewed by others
References
Ventosa A (2006) Unusual micro-organisms from unusual habitats: hypersaline environments. In: Logan NA, Lappin-Scott HM, Oyston PCF (eds) Prokaryotic diversity: mechanisms and significance: published for the Society for General Microbiology, Symposium no. 66, Cambridge University Press, Cambridge, pp 223–253. https://doi.org/10.1017/CBO9780511754913.015
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271. https://doi.org/10.1099/ijs.0.038653-0
Oren A, Ventosa A, Kamekura M (2017) Halobacteria. Bergey’s manual of systematics of archaea and bacteria. Wiley, Hoboken, pp 1–5. https://doi.org/10.1002/9781118960608.cbm00026.pub2
Gupta RS, Naushad S, Baker S (2015) Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol 65:1050–1069. https://doi.org/10.1099/ijs.0.070136-0
Parte AC (2018) LPSN - List of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int J Syst Evol Microbiol 68(6):1825–1829. https://doi.org/10.1099/ijsem.0.002786
Hezayen FF, Tindall BJ, Steinbüchel A, Rehm BHA (2002) Characterization of a novel halophilic archaeon, Halobiforma haloterrestris gen. nov., sp. nov., and transfer of Natronobacterium nitratireducens to Halobiforma nitratireducens comb. nov. Int J Syst Evol Microbiol 52:2271–2280. https://doi.org/10.1099/00207713-52-6-2271
Castillo AM, Gutiérrez MC, Kamekura M et al (2006) Halostagnicola larsenii gen. nov., sp. nov., an extremely halophilic archaeon from a saline lake in Inner Mongolia, China. Int J Syst Evol Microbiol 56:1519–1524. https://doi.org/10.1099/00207713-52-6-2271
Gutiérrez MC, Castillo AM, Kamekura M et al (2007) Halopiger xanaduensis gen. nov., sp. nov., an extremely halophilic archaeon isolated from saline Lake Shangmatala in Inner Mongolia, China. Int J Syst Evol Microbiol 57:1402–1407. https://doi.org/10.1099/ijs.0.65001-0
Ding JY, Chen SC, Lai MC, Liao TL (2017) Haloterrigena mahii sp. Nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 67(5):1333–1338. https://doi.org/10.1099/ijsem.0.001811
Nagaoka S, Minegishi H, Echigo A, Usami R (2010) Halostagnicola kamekurae sp. nov., an extremely halophilic archaeon from solar salt. Int J Syst Evol Microbiol 60:2828–2831. https://doi.org/10.1099/ijs.0.014449-0
Roh SW, Do NY, Chang HW et al (2009) Haloterrigena jeotgali sp. nov., an extremely halophilic archaeon from salt-fermented food. Int J Syst Evol Microbiol 59:2359–2363. https://doi.org/10.1099/ijs.0.008243-0
Albuquerque L, Taborda M, La Cono V et al (2012) Natrinema salaciae sp. nov., a halophilic archaeon isolated from the deep, hypersaline anoxic Lake Medee in the Eastern Mediterranean Sea. Syst Appl Microbiol 35(6):368–373. https://doi.org/10.1016/j.syapm.2012.06.005
McGenity TJ, Gemmell RT, Grant WD (1998) Proposal of a new halobacterial genus Natrinema gen. nov., with two species Natrinema pellirubrum nom. nov. and Natrinema pallidum nom. nov. Int J Syst Bacteriol 48:1187–1196. https://doi.org/10.1099/00207713-48-4-1187
Nagaoka S, Minegishi H, Echigo A et al (2011) Halostagnicola alkaliphila sp. nov., an alkaliphilic haloarchaeon from commercial rock salt. Int J Syst Evol Microbiol 61:1149–1152. https://doi.org/10.1099/ijs.0.023119-0
Tindall BJ, Ross HNM, Grant WD (1984) Natronobacterium gen. nov. and Natronococcus gen. nov., two new genera of haloalkaliphilic archaebacteria. Syst Appl Microbiol 5(1):41–57. https://doi.org/10.1016/S0723-2020(84)80050-8
Kanai H, Kobayashi T, Aono R, Kudo T (1995) Natronococcus amylolyticus sp. nov., a haloalkaliphilic archaeon. Int J Syst Bacteriol 45(4):762–766. https://doi.org/10.1099/00207713-45-4-762
Roh SW, Do NY, Chang HW et al (2007) Natronococcus jeotgali sp. nov., a halophilic archaeon isolated from shrimp jeotgal, a traditional fermented seafood from Korea. Int J Syst Evol Microbiol 57(Pt 9):2129–2131. https://doi.org/10.1099/ijs.0.65120-0
Corral P, Gutiérrez MC, Castillo AM et al (2013) Natronococcus roseus sp. nov., a haloalkaliphilic archaeon from a hypersaline lake. Int J Syst Evol Microbiol 63(Pt 1):104–108. https://doi.org/10.1099/ijs.0.036558-0
Kajale S, Deshpande N, Shouche Y, Sharma A (2020) Cultivation of diverse microorganisms from hypersaline lake and impact of delay in sample processing on cell viability. Curr Microbiol 77:716–721. https://doi.org/10.1007/s00284-019-01857-8
Sharma A, Shouche Y (2014) Microbial culture collection (MCC) and international depositary authority (IDA) at National Centre for Cell Science, Pune. Indian J Microbiol 54:129. https://doi.org/10.1007/s12088-014-0447-y
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238. https://doi.org/10.1099/00207713-47-1-233
Sharma A, Pandey A, Shouche YS et al (2009) Characterization and identification of Geobacillus spp. isolated from Soldhar hot spring site of Garhwal Himalaya, India. J Basic Microbiol 49:187–194. https://doi.org/10.1002/jobm.200800194
Kajale S, Deshpande N, Pali S et al (2020) Natrialba swarupiae sp. nov., a halophilic archaeon isolated from a hypersaline lake in India. Int J Syst Evol Microbiol 70(3):1876–1881. https://doi.org/10.1099/ijsem.0.003986
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67(5):1613–1617. https://doi.org/10.1099/ijsem.0.001755
Sharma A, Dhar SK, Prakash O et al (2014) Description of Domibacillus indicus sp. nov., isolated from ocean sediments and emended description of the genus Domibacillus. Int J Syst Evol Microbiol 64:3010–3015. https://doi.org/10.1099/ijs.0.064295-0
Wattam AR, Davis JJ, Assaf R et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45(D1):D535–D542. https://doi.org/10.1093/nar/gkw1017
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Parks DH, Imelfort M, Skennerton CT et al (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Yoon SH, Ha SM, Lim J et al (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Rodriguez-R LM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe Mag 9:111–118. https://doi.org/10.1128/MICROBE.9.111.1
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y) 39:783–791. https://doi.org/10.2307/2408678
Na SI, Kim YO, Yoon SH et al (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285. https://doi.org/10.1007/s12275-018-8014-6
Chun J, Oren A, Ventosa A et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Oren A (2012) The function of gas vesicles in halophilic archaea and bacteria: theories and experimental evidence. Life 3(1):1–20. https://doi.org/10.3390/life3010001
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485. https://doi.org/10.1128/jb.70.4.484-485.1955
Cui HL, Sun FF, Gao X et al (2010) Haladaptatus litoreus sp. nov., an extremely halophilic archaeon from a marine solar saltern, and emended description of the genus Haladaptatus. Int J Syst Evol Microbiol 60(5):1085–1089. https://doi.org/10.1099/ijs.0.015933-0
Sharma A, Jani K, Feng GD et al (2018) Subsaxibacter sediminis sp. nov., isolated from arctic glacial sediment and emended description of the genus Subsaxibacter. Int J Syst Evol Microbiol 68:1678–1682. https://doi.org/10.1099/ijsem.0.002729
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. https://doi.org/10.1139/o59-099
Card GL (1973) Metabolism of phosphatidylglycerol, phosphatidylethanolamine, and cardiolipin of Bacillus stearothermophilus. J Bacteriol 114:1125–1137. https://doi.org/10.1128/jb.114.3.1125-1137.1973
Soto CY, Cama M, Gibert I, Luquin M (2000) Application of an easy and reliable method for sulfolipid-I detection in the study of its distribution in Mycobacterium tuberculosis strains. FEMS Microbiol Lett 187:103–107. https://doi.org/10.1016/S0378-1097(00)00183-X
Acknowledgements
We are grateful to Prof. Aharon Oren and Prof. Bernhard Schink for the Latin etymology and species epithet. We are grateful to Kunal Jani for his help in sample collection.
Funding
This work was supported by the Department of Biotechnology (DBT), Government of India (Grant No. BT/Coord.II/01/03/2016), under the project ‘Establishment of National Centre for Microbial Resource’.
Author information
Authors and Affiliations
Contributions
SK isolated the strain LS1_41T, performed phenotypic, biochemical, genomic and phylogenetic analysis and wrote the manuscript; TL performed the polar lipid analysis; ND, YS and AS supervised the study; AS designed study and revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This study was not performed on human or animals, therefore no ethical approval is required.
Consent for Publication
All authors approve to submit and publication to the journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kajale, S., Deshpande, N., Lodha, T. et al. Natronococcus pandeyae sp. nov., a Novel Haloarchaeon from Sambhar Salt Lake. Curr Microbiol 79, 51 (2022). https://doi.org/10.1007/s00284-021-02740-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-021-02740-1