Abstract
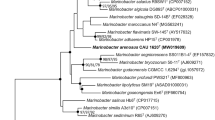
A Gram stain-negative, aerobic, motile, and rod-shaped bacterium designated 176SS1-4T was isolated from the salt-field sea water. Strain 176SS1-4T grew well at 25 °C on marine agar. The taxonomic affiliation of the novel isolate was identified using the polyphasic approach. By comparative 16S rRNA gene sequence, it could be shown that strain 176SS1-4T branches within the family Rhodobacteraceae and class Alphaproteobacteria and is related to Histidinibacterium lentulum B17T (96.0% sequence similarity). The digital DNA–DNA hybridization and average nucleotide identity between strain 176SS1-4T and H. lentulum B17T was 19.2% and 77%. The genome comprises of 3,905,029 bp with a G + C content of 67.7 mol. Ubiquinone 10 (Q-10) was the major respiratory quinone. The major fatty acids were summed feature 8 (comprising C18:1 ω7c and/or C18:1 ω6c), C16:0, C19:0 cyclo ω8c, C18:0, and C18:1 ω6c 11-methyl and the major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylcholine. On the basis of the genotypic and phenotypic characteristics, strain 176SS1-4T can be placed as a novel species within the genus Histidinibacterium; the name Histidinibacterium aquaticum sp. nov. has been proposed, with type strain 176SS1-4T (= KACC 19891T = LMG 31032T).

Similar content being viewed by others
References
Oren A (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4:2
Ventosa A, de la Haba RR, Sánchez-Porro C, Papke RT (2015) Microbial diversity of hypersaline environments: a metagenomic approach. Curr Opin Microbiol 25:80–87
Pastor JM, Bernal V, Salvador M, Argandoña M, Vargas C, Csonka L, Sevilla A, Iborra JL, Nieto JJ, Cánovas M (2013) Role of central metabolism in the osmoadaptation of the halophilic bacterium Chromohalobacter salexigens. J Biol Chem 288:17769–17781
Pflüger K, Baumann S, Gottschalk G, Lin W, Santos H, Müller V (2003) Lysine-2, 3-aminomutase and ß-lysine acetyltransferase genes of methanogenic Archaea are salt induced and are essential for the biosynthesis of NΕ-acetyl-ß-lysine and growth at high salinity. Appl Environ Microbiol 69:6047–6055
Saum R, Mingote A, Santos H, Müller V (2009) A novel limb in the osmoregulatory network of Methanosarcina mazei Go¨1:Ne-acetyl-ß-lysine can be substituted by glutamate and alanine. Environ Microbiol 11:1056–1065
Kamekura M (1986) Production and function of enzymes from eubacterial halophiles. FEMS Microbiol Rev 39:145–150
Ben-Amotz A, Avron M (1989) The biotechnology of mass culturing Dunaliella for products of commercial interest. In: Cresswell RC, Rees TAV, Shah N (eds) Algal and cyanobacteiral Biotechnology. Longman Scientific and Technical Press, London, pp 91–114
Wang G, Xu S, Su H, Chen B, Liang J, Huang W, Wang Y, Yu K (2019) Histidinibacterium lentulum gen. nov., sp. nov., a marine bacterium from the culture broth of marine microalga Picochlorum sp. Int J Syst Evol Microbiol 69(2):578–583
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Perry LB (1973) Gliding motility in some non-spreading flexibacteria. J Appl Bacteriol 36:227–232
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Pearson Education Inc, San Francisco, CA
Atlas RM (1993) Handbook of microbiological media. CRC Press, Boca Raton, FL
Ten LN, Im WT, Kim MK, Kang MS, Lee ST (2004) Development of a plate technique for screening of polysaccharide-degrading microorganisms by using a mixture of insoluble chromogenic substrates. J Microbiol Methods 56:375–382
Kim JK, Kang MS, Park SC, Kim KM, Choi K, Yoon MH, Im WT (2015) Sphingosinicella ginsenosidimutans sp. nov., with ginsenoside converting activity. J Microbiol 53:435–441
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge, NY
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark, DE
Li R (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A largescale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Li FN, Liao SL, Liu SW, Jin T, Sun CH (2019) Aeromicrobium endophyticum sp. nov an endophytic actinobacterium isolated from reed (Phragmites australis). J Microbiol. https://doi.org/10.1007/s12275-019-8705-7
Acknowledgements
This research was supported by National Marine Biodiversity Institute of Korea (2021M01100) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A3046479).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The NCBI GenBank accession number for the 16S rRNA gene sequence and genome sequence of strain 176SS1-4T are MK040417 and VYQE01000000.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2021_2688_MOESM2_ESM.pptx
Supplementary Fig. S2. Two-dimensional TLC of polar lipids of strain 176SS1-4T. The following spray reagents were used for detection: (A) molybdenum blue (Sigma) (for phospholipids); (B) 5% ethanolic molybdophosphoric acid (Sigma) (for total lipids); (C) ninhydrin (Sigma) (for aminolipids). Abbreviations: DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; PC, phosphatidylcoline; AL, aminolipids (AL1, AL2); L, unidentified polar lipids (L1- L3) (PPTX 334 kb)
Rights and permissions
About this article
Cite this article
Lee, KH., Park, JS. Histidinibacterium aquaticum sp. nov., Isolated from Salt-Field Sea Water. Curr Microbiol 79, 67 (2022). https://doi.org/10.1007/s00284-021-02688-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-021-02688-2