Abstract
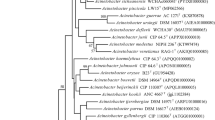
A Gram stain negative, motile, non-spore-forming, rod-shaped, strictly aerobic, beige-pigmented bacterium, designated strain BO-7T, was isolated from soil of cattle farm, in Seosan, Republic of Korea. On the basis of 16S rRNA gene sequencing, strain BO-7T clustered with species of the genus Ochrobactrum and appeared closely related to O. haematophilum CCUG 38531T (98.9%), O. daejeonense KCTC 22458T (98.1%), O. rhizosphaerae DSM 19824T (98.1%), O. pituitosum DSM 22207T (98.0%), and O. pecoris DSM 23868T (98.0%). The digital DNA-DNA hybridization and average nucleotide identity between strain BO-7T and the closely related strains were 21.9–39.1%, 78.5–89.5%, respectively, indicating that BO-7T is a novel species of the genus Ochrobactrum. The DNA G + C content of the genomic DNA was 57.1 mol%, and ubiquinone Q-10 was the predominant respiratory quinone. The polar lipids consisted of phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, phosphatidylmonomethyl-ethanolamine, di-phosphatidylglycerol, the major polyamines were spermidine, putrescine, and sym-homospermidine. The major cellular fatty acids (> 5%) were C16:0, C19:0 cycle ω7c, and C18:1ω7c and/or C18:1ω6c (summed feature 8). ANI calculation, digital DNA-DNA hybridization, physiological and biochemical characteristics indicated that strain BO-7T represents a novel species of the genus Ochrobactrum, for which the name Ochrobactrum soli sp. nov. is proposed. The type strain is BO-7T (= KACC 19676T = LMG 30809T).


Similar content being viewed by others
References
Holmes B, Popoff M, Kiredjian M, Kersters K (1988) Ochrobactrum anthropi gen. nov., sp. nov. from human clinical specimens and previously known as group Vd. Int J Syst Bacteriol 38:406–416
Lebuhn M, Achouak W, Schloter M, Berge O, Meier H, Barakat M, Hartmann A, Heulin T (2000) Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int J Syst Evol Microbiol 50:2207–2223
Huber B, Scholz HC, Kämpfer P, Falsen E, Langer S (2010) Ochrobactrum pituitosum sp. nov., isolated from an industrial environment. Int J Syst Evol Microbiol 60:321–326
Kämpfer P, Sessitsch A, Schloter M, Huber B, Busse HJ, Scholz HC (2008) Ochrobactrum rhizosphaerae sp. nov. and Ochrobactrum thiophenivorans sp. nov., isolated from the environment. Int J Syst Evol Microbiol 58:1426–1431
Kämpfer P, Huber B, Busse HJ, Scholz HC, Tomaso H, Hotzel H, Melzer F (2011) Ochrobactrum pecoris sp. nov., isolated from farm animals. Int J Syst Evol Microbiol 61:2278–2283
Li L, Li YQ, Jiang Z, Gao R, Nimaichand S, Duan YQ, Egamberdieva D, Chen W, Li WJ (2016) Ochrobactrum endophyticum sp. nov., isolated from roots of Glycyrrhiza uralensis. Arch Microbiol 198:171–179
Woo SG, Ten LN, Park J, Lee J (2011) Ochrobactrum daejeonense sp. nov., a nitrate-reducing bacterium isolated from sludge of a leachate treatment plant. Int J Syst Evol Microbiol 61:2690–2696
Imran A, Hafeez FY, Frühling A, Schumann P, Malik KA, Stackebrandt E (2010) Ochrobactrum ciceri sp. nov., isolated from nodules of Cicer arietinum. Int J Syst Evol Microbiol 60:1548–1553
Gazolla VC, Hayashi SF, Ambrosini A, Brito LB, Kayser VL, Passaglia LMP (2019) Reclassification of Ochrobactrum lupini as a later heterotypic synonym of Ochrobactrum anthropi based on whole-genome sequence analysis. Int J Syst Evol Microbiol 96:2312–2314
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Perry LB (1973) Gliding motility in some non-spreading flexibacteria. J Appl Bacteriol 36:227–232
Weon HY, Kim BY, Joa JH, Son JA, Song MH, Kwon SW, Go SJ, Yoon SH (2008) Methylobacterium iners sp. nov. and Methylobacterium aerolatum sp. nov., isolated from air samples in Korea. Int J Syst Evol Microbiol 58:93–96
Cappuccino JG, Sherman N (2002) Microbiology: a laboratory manual, 6th edn. Pearson Education Inc, California
Atlas RM (1993) Handbook of microbiological media. CRC Press, Boca Raton, Florida, USA
Cowan ST, Steel KJ (1974) Manual for the identification of medical bacteria. Cambridge University Press, Cambridge
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (ed) Nucleic acid techniques in bacterial systematics. Wiley, New York.
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Kämpfer P, Scholz HC, Huber B, Falsen E, Busse HJ (2007) Ochrobactrum haematophilum sp. nov. and Ochrobactrum seudogrignonense sp. nov., isolated from human clinical specimens. Int J Syst Evol Microbiol 57:2513–2518
Li R (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A largescale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Li FN, Liao SL, Liu SW, Jin T, Sun CH (2019) Aeromicrobium endophyticum sp. nov., an endophytic actinobacterium isolated from reed (Phragmites australis). J Microbiol https://doi.org/10.1007/s12275-019-8705-7
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469
Busse J, Auling G (1988) Polyamine pattern as a chemotaxonomic marker within the Proteobacteria. Syst Appl Microbiol 11:1–8
Schenkel E, Berlaimont V, Dubois J, Helson CM, Hanocq M (1995) Improved high-performance liquid chromatographic method for the determination of polyamines as their benzoylated derivatives: application to P388 cancer cells. J Chromatogr B 668:189–197
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark.
Alanjary M, Steinke K, Ziemert N (2019) AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res 47:W276–W282
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Acknowledgements
This research was supported by the project on survey and excavation of Korean indigenous species of the National Institute of Biological Resources (NIBR) and by a grant from the Korea Research Institute of Bioscience & Biotechnology (KRIBB) Research Initiative Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The NCBI GenBank Accession Number for the 16S rRNA gene sequence and genome sequence of strain BO-7T are MH094651 and RCHH00000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2020_1882_MOESM2_ESM.pptx
Supplementary file2 Phylogenetic tree (constructed with neighbour-joining method) showing the relationships of strain BO-7T with other related species of the genus Ochrobactrum. Filled circles indicate that the corresponding nodes were also recovered in trees generated with the maximum-likelihood and maximum-parsimony algorithm. Bootstrap values (expressed as percentages of 1,000 replications) greater than 60% are shown at the branch points (PPTX 49 kb)
284_2020_1882_MOESM3_ESM.pptx
Supplementary file3 The phylogenetic tree was constructed from a comparative analysis of whole genome sequences showing the relationships of strain BO-7T with other related species of the family Brucellaceae. This tree was constructed via Automated Multi-Locus Species Tree online web 36 server, and with Mega 7 program using the aligned sequences of Automated Multi-Locus Species 36 analysis. Bootstrap values are 30 shown at the branch points. The Bar represents 0.1 substitutions per nucleotide position (PPTX 56 kb)
284_2020_1882_MOESM4_ESM.pptx
Supplementary file4 Two-dimensional thin-layer chromatography of the total polar lipids of strain BO-7T. Chloroform-methanol-water (65:25:4, by vol.) was used in the first direction, followed by chloroform-acetic acid-methanol-water (40:7.5:6:2, by vol.) in the second direction. The following spray reagents were used for detection: (a) 5% ethanoic molybdophosphoric acid (for total lipids); (b) molybdenum blue (Sigma) (for phosphorus-containing polar lipids); (c) 2% ninhydrin (for amino lipids); (d) Dragendorff reagent (for choline lipid). PE, phosphatidylethanolamine; PME, phosphatidylmonomethylethanolamine; PG, phosphatidylglycerol; DPG, diphosphatidylglycerol; PC, phosphatidylcholine; AL1–2, unidentified amino-lipids; L1, unidentified polar lipid (PPTX 1741 kb)
Rights and permissions
About this article
Cite this article
Choi, GM., Kim, K.M., Yun, CS. et al. Ochrobactrum soli sp. nov., Isolated from a Korean Cattle Farm. Curr Microbiol 77, 1104–1110 (2020). https://doi.org/10.1007/s00284-020-01882-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01882-y