Abstract
Ducks play an important role in transmitting and maintaining mammalian viruses in nature, and are a reservoir host of many animal viruses. We analyzed the fecal virome of four strains (A, B, C, and D) of ducks living in isolation by using metagenomic analysis. The feces of the ducks tested contained 18 animal virus families. The percentage values of RNA virus reads, compared to the total animal virus reads in each of the four strains were 96.96% (A), 97.30% (B), 98.01 (C), and 67.49% (D), and were mainly from Orthomyxoviridae, Mimiviridae, Bunyaviridae, Picobirnaviridae, and Reoviridae. Meanwhile, the minority of DNA virus reads were related to Herpesviridae, Adenoviridae, Iridoviridae, and other, low abundance viral families. The percentage values of Orthomyxoviridae, Mimiviridae, Bunyaviridae, Picobirnaviridae, and Herpesviridae reads were not significantly different among strains A, B, and C; however, there were marked differences in the abundance of these reads in strain D. In summary, this study provides an unbiased examination of the viral diversity in the feces of four strains of ducks in specific-pathogen-free periods, and highlights the variation in the percentage of viral families present. These results can be used as a reference for detecting duck viral pathogens and predicting zoonotic potential.
Similar content being viewed by others
Introduction
Ducks are aquatic birds belonging to the order Anseriformes [1]. China is one of the most prominent countries for duck breeding in world, having the highest duck slaughter rates and breeding stocks. Domestic ducks and waterfowl live side by side in the rice fields and wetlands in China, and the common-living environment provides opportunity for contact between wild and domestic birds [2]. The ducks are a reservoir host of mammalian viruses in nature. Many studies indicate that ducks and geese are more responsible for virus transmission over large geographic distances than gulls and shorebirds [3, 4]. Many reports suggest that low pathogenic avian influenza (LPAI) strains are more frequently isolated from ducks than gulls and shorebirds [3, 5]. This demonstrates that ducks play an important role in amplifying and hosting a range of mammalian viruses [6, 7].
The traditional methods for the study of viral diseases are limited and have many shortcomings, such as their inability to replicate viruses in cell culture, difficulty in detecting unknown viral sequences by PCR, and the lack of cross-reacting antibodies for known viruses [8]. With the development and use of viral metagenomic analyses, many of these limitations and shortcomings can now be avoided. These methods have been used since their inception to identify the unbiased viral diversity from the digestive tract of humans [8], bats [9], turkey [10], pigs [11], pigeons [12], and ducks [13].
In this study, we used viral metagenomics to explore a new method to detect known duck viruses and obtain the viral spectrum of ducks in specific-pathogen-free (SPF) revolution periods while living in isolation. We described the duck viral families identified, which included Orthomyxoviridae, Herpesviridae, Picobirnaviridae, Reoviridae, Adenoviridae, Flaviviridae, Poxviridae, Circoviridae, and other uncharacterized duck viral families, excluding insects, plants, and phage viral reads. These results provided a baseline fecal virome for ducks in isolation, which could be used as a reference for purifying duck viral pathogens in the SPF revolution period, and in predicting future viral disease outbreaks.
Materials and Methods
Duck Feces Samples
Jinding ducks, known as mallards, belong to the sheldrakes and are laying ducks in Jiangsu province, China. The Jinding ducks are known for high laying rate, therefore, we introduced progenitor Jinding eggs of four strains of ducks (A, B, C, and D) from Jiangsu’s Fengda waterfowl breeding field of the national waterfowl germplasm resources gene pool. The ducks were divided into four strains (A, B, C, and D) based on genetic background. In order to obtain the SPF ducks, the ducklings lived in isolation from hatching. We detected the viral pathogens [duck hepatitis A virus (DHAV), duck plague virus (DPV), avian leukosis virus (ALV), avian reovirus (ARV), goose/duck parvovirus (GPV), duck circovirus (DuCV), duck Tembusu virus (DTMUV), and Newcastle disease virus (NDV)] and antibodies (avian adenovirus group III, NDV, H5, H7, and H9 subtypes AIV) by using polymerase chain reaction (PCR) and hemagglutination inhibition tests once a month in the first 3 months, then once every 2 months. Once any virus pathogens were detected in the ducks, they were eliminated immediately. Fecal specimens from four strains were collected in December 2015. Three fecal samples were taken from ducks of each strain, and three fecal samples from the same strain were mixed into a single sample. The sample was immediately suspended in phosphate buffer saline (PBS).
The stool suspension was centrifuged at 12,000×g at 4 °C and the pellet was discarded. The supernatant was transferred to a new tube and centrifuged for a second time. The supernatant was filtered through a 0.22 µm filter, in order to remove the nucleic acid not in viral capsid, and we added DNaseI (Roche Basel, Switzerland) and RNaseH (Toyobo, Tokyo, Japan) to the filtrates [8]. Viral DNA was isolated according to the traditional phenol/chloroform extraction method, and viral RNA was extracted using RNAiso plus (Takara, Dalian, China) [14, 15]. The extracted RNA was reverse-transcribed using SISPA (Sequence-independent, single primer amplification), with the K-8N primer (GACCATCTAGCGACCTCC ACNNNNNNNN) described previously [16,17,18]. The dsDNA and viral DNA were mixed and detected using the Qubit dsDNA HS assay (Invitrogen, Carlsbad, CA, USA). After product inspection, the sample was sheared into random 300 bp fragments and prepared for sequencing using PE150, Illumina HiSeq 4000 [19].
Bioinformatics
The raw data were pretreated by removing the low-quality data, which accounted for more than 40% of the reads including bases reaching a certain proportion of reads; overlap between adapters above a certain threshold value; and if the sample was polluted by the host the results were compared with the host database and probable host reads filtered out. The pretreated sample (Clean Data) was blasted with the NCBI viral genome database, NT viruses database, and ACLAME database by setting a threshold e value less than e−3 [9]. The reads of clean data were classified into family, genus, and species.
Verification of AIV and DPV
We used the national standard M gene primers (MF: TTCTAACCGAGGTCGAAAC; MR: AAGCGTCTACGCTGCAGTCC) to detect AIV, and purified PCR product, then sequenced, and analyzed sequence of the product. DPV was detected using PCR with GBF and GBR primers (GBF: GAGCGTATTTAGTAGAAACTGC; GBR: TGAATGTTGTGATTGTTC).
Results
Viral Sequences Overview
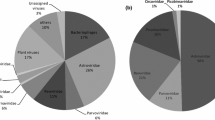
Stool samples were collected from four strains (A, B, C, and D) of ducks in isolation. Viral nucleic acids were enriched and purified by concentration and filtration, then sequenced, generating >61,139,452 reads. Sequence reads for each strain were classified based on Blastx scores (E ≤ e−3). Some 87.28, 82.85, 84.53, and 59.54% of the reads from the A, B, C, and D strains were not annotated to any viral sequences from the databases, respectively (Fig. 1). The virus reads of the D strain was more than the other three strains.
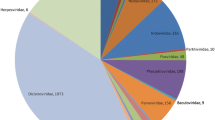
The most abundant viral families in each of the four duck strains were different. The majority of virus family reads in strain D belonged to animal viral families, while the phage viral families dominated in strain C. In strain B, the most abundant viral families were Inoviridae and Caulimoviridae, meanwhile, the most prevalent viral families from strain A belonged to DNA viruses (Fig. 2). The annotated reads mainly belonged to 34 viral families as in the four strains (each virus family reads of three groups >50). In addition to phage reads, there were 27 viral families. Some [1.31% (A), 0.91% (B), 2.62% (C), and 0.20% (D)] of the viral reads were not classified as any known viral families (Fig. 3). The plant viral reads were 0.75% (A), 0.39% (B), 0.27% (C), and 0.11% (D) of the total viral reads of each strain, while 0.23% (A), 0.14% (B), 0.40% (C), and 0.04% (D) of each strain were contributed by insect reads (Fig. 3). These results indicated that the composition of unclassified viral reads, plant viral reads, and insect viral reads were largely unchanged among the four duck strains, compared to the large differences in phage and animal viral reads between the strains.
The phage viral reads were 37.23% (A), 16.99% (B), 29.21% (C), and 1.59% (D) of the total viral reads of each strain (Fig. 3). The fecal samples of the four strains (A, B, C, and D) mainly contained major double-stranded DNA myoviruses, siphoviruses, and podoviruses, and single-stranded DNA inoviruses and microviruses. The phage reads in the four strains had different components. The percentage of phage reads in strain D was the lowest among the four strains (Fig. 4). The bacteriophage-like reads of the four strains of duck feces were dominated by double-stranded DNA phage reads (Fig. 4). However, the Myovirus reads from strains A and D were twice as prevalent as in the B and C strains. The percentage of Inoviridae reads in strain B was 60–120 times higher than in the A, C, and D strains (Fig. 4).
Comparison of Animal Viral Reads in Different Duck Strains
The percentage values of eukaryotic animal virus reads in the four duck strains (A, B, C, and D) viral sequence reads were 60.48, 81.57, 67.50, and 98.06%, respectively (Fig. 3). All such reads comprised 18 viral families. Next, the fecal animal viral reads of the A, B, C, and D strains were compared. A total of 96.96% (A), 97.30% (B), 98.01% (C), and 67.49% (D) of animal viral reads in each strain were related to RNA viruses from Orthomyxoviridae, Mimiviridae, Bunyaviridae, Picobirnaviridae, Reoviridae, and five other viral families (Fig. 5). The percentage values of Orthomyxoviridae, Mimiviridae, Bunyaviridae reads compared to the total animal virus reads were lowest for strain D (64.91, 0.04, and 0.03%, respectively) and highest for strain A (95.59, 0.43, and 0.14%, respectively), B (96.48, 0.20, and 0.10%), and C (96.93, 0.28, and 0.18%). However, the percentage values of Picobirnaviridae reads, compared to the total animal virus reads were 0.13% (intermediate), 0.03, 0.01% (lowest), and 2.26% (highest) for strains A, B, C, and D, respectively (Fig. 5). The minority of viral reads matched to DNA viruses, including Herpesviridae, Adenoviridae, Iridoviridae, as well as five other viral families (Fig. 5). There was no marked difference in the percentage of Adenoviridae and Iridoviridae among strains A, B, C, and D. A total of 2.23% (A), 2.10% (B), and 1.45% (C) of the total animal virus reads were from Herpesviridae, which was much more abundant in strain D (32.24%) (Fig. 5).
Validation of AIV and DPV
Orthomyxoviridae and Herpesviridae virus are important pathogens for healthy duck breeding. In our study, Orthomyxoviridae and Herpesviridae reads were the most abundant. AIV is a segmented negative-strand RNA virus enveloped in capsid protein and belongs to the Orthomyxoviridae family [20, 21]. Aquatic birds are a migration reservoir host of LPAI, which is occasionally transmitted from aquatic birds to other animal hosts, including mammals. Aquatic domestic poultry also play an important role in the spread and maintenance of viruses in nature. There were 1,198,204, 2,109,547, 1,341,972, and 4,187,452 AIV reads of the A, B, C, and D strains in the annotated sequence reads, respectively. According to the national standard M gene PCR primers, the PCR amplified product of all four strains is 229 bp. When sequenced and blasted, the sequence had 98% identity with the AIV sequence from the NCBI database.
DPV belongs to the family Herpesviridae and its genome is composed of double-stranded DNA and is 158,091 bp in length [22]. DPV causes acute contagious infections among ducks, geese, and swans, and is known as duck virus enteritis (DVE) and duck plague (DP). DVE causes high morbidity and mortality, which leads to enormous economic losses in waterfowl breeding [23]. Herpesviridae reads dominated among the DNA animal virus reads; however, identification of DEV reads in four strains was rare. No amplification products were detected. These results indicated that the metagenomic analysis was reliable.
Discussion
Migratory aquatic birds are the intermediate hosts of many viruses. Domestic ducks and waterfowl live side by side in the rice fields and wetlands in China. The way in which ducks feed provides the conditions for spreading viruses between wild birds and ducks [2]. Ducks play an important role in spreading and maintaining viruses in nature. We described here the contents of viral sequence reads of four strains (A, B, C, and D) of ducks in an SPF evolution period.
The four strains contained the five phage viruses as follows: myoviruses, siphoviruses, podoviruses, inoviruses, and microviruses. This was consistent with the previously reported phage composition of ducks, human, pig, and equine feces by metagenomic analysis [11, 13, 24,25,26]. A high percentage (99.43, 69.17, 95, 97.68%) of phage reads was double-stranded DNA viruses and belonged to the order Caudovirales, while the B strain had a higher percentage of inovirus reads than the A, C, or D strains. The differences between the four strains reflected subtle difference among intestinal flora environments. However, the similar phage contents of the four strains suggested that they have almost the same composition of gut bacteria.
Duck stool and blood samples were detected seven times by PCR before metagenomic analysis. Once AIV, DHAV, DPV, ALV, ARV, avian adenovirus group III, GPV, DuCV, DTMUV, or DNV were detected, the ducks were eliminated. However, all four strains included about 300 species of virus, showing a high level of mixed infections. This high level of coinfections may be an overestimate. If the reads are aligned, assembled, and analyzed, the number of virus sequence reads from a sample may decrease. AIV, DHAV, and ALV reads were found in all four strains. One and two GPV reads were detected in the A and C strains, respectively. Duck circovirus reads were detected in the B strain. Anatid herpesvirus 1, known as DPV, Fowlpox virus, Canarypox virus, and pigeonpox virus reads related to duck were found and relatively scarce, and those viral reads were identified in Indian ducks feces [13]. The result suggested that the sensitivity of PCR was not high; we should adopt a variety of methods to identify pathogens in samples. The fecal viral reads of ducks contained avian Coronavirus reads; however, we did not detect this viral pathogen in the ducks in our study. This provided a baseline reference for purifying the ducks in isolation.
The fecal samples contained plant and insect viral reads, including Geminiviridae, Partitiviridae, Phycodnaviridae, Bunyaviridae, Caulimoviridae, Alphanodavirus, Iridoviridae, and Baculoviridae, reflecting the ducks’ diet. The feces of sea lions contained insect virus reads [27], and the plant and insect virus reads were also detected from human feces [28, 29]. The plant and insect virus reads were different from the previous metagenomic analysis of duck gut [13], indicating that the diet and social habitation of Jinding ducks and Indian wild ducks were different. The majority of virus reads of fecal samples were animal viruses, including AIV, cercopithecine herpesvirus, human picobirnavirus, bluetongue virus, murine mastadenovirus A, Alphapapillomavirus 9, and human endogenous retrovirus, which could infect mammals. AIV also infects birds. Ducks are aquatic birds that live in the same environment as wild birds and act as intermediate hosts between mammals and wild birds. Thus, the mammalian viral reads from duck feces provides a wake-up call for the potential for anthropozoonosis.
The four strains had similar compositions of animal viruses; however, the percentage of per viral family reads exhibited differences among the four strains. The percentage of annotated viral reads, compared to the total reads, and the percentage of animal virus reads in the total viral reads of strain D were significantly higher than the A, B, and C strains. However, the percentage of RNA virus reads in the total animal virus reads was lowest for strain D, compared to the other strains. Meanwhile, the percentage of Picobirnaviridae and Herpesviridae reads was highest for strain D, while the percentage values of Orthomyxoviridae, Mimiviridae, Bunyaviridae reads in strain A, strain B, and C strain were markedly higher than in strain D. These results suggested that the D strain ducks were more susceptible to viruses. The susceptibility and adsorption of the C strain may be very low for viruses. However, the difference in abundance of each viral family among the four duck strains reflected that the susceptibility of different strains for one viral family may be different. The four strains were divided based on their genetic background. The division genes of these strains may be key factors for adsorption, susceptibility, and innate immune response to viruses. The differences that occur among the strains may be related to susceptibility genes and the innate immune-related host genes [30]. However, Pekin, Muscovy, and mallard ducks infected with an H5N1 HPAI showed a systemic infection with high mortality. Muscovy ducks produced more severe clinical symptoms and had a poor response to vaccination. Nevertheless, the expression of innate immune-related genes was similar in the spleens of ducks infected with viruses [31]. In this study, we did not detect the expression of innate immune-related genes from the four duck strains. The expression of innate immune-related genes of the four duck strains will be examined and compared in future studies.
In summary, this study provided an unbiased overview of the fecal virome of ducks and compared the similarities and differences of viral family reads among four duck strains. Sequencing of the fecal virome of ducks by metagenomic analysis should provide a useful method to further examine a range of mammalian viruses of SPF ducks. Characterizing the feces virome of four genetically distinct duck strains provided a reference for purifying duck viral pathogens and facilitating improvements in animal health and production.
References
Firdous AD, Maya S, Massarat K, Baba MA (2016) Developmental ossification sequences of the appendicular and axial skeleton in Kuttanad duck embryos (Anas platyrhynchos domesticus). Open Vet J 6(1):1–5
Hill NJ, Runstadler JA (2016) A bird’s eye view of influenza A virus transmission: challenges with characterizing both sides of a co-evolutionary dynamic. Integr Comp Biol 56(2):304–316
Chen GQ, Zhuang QY, Wang KC, Liu S, Shao JZ, Jiang WM, Hou GY, Li JP, Yu JM, Li YP, Chen JM (2013) Identification and survey of a novel avian coronavirus in ducks. PLoS ONE 8(8):e72918
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56(1):152–179
Costa TP, Brown JD, Howerth EW, Stallknecht DE (2011) Variation in viral shedding patterns between different wild bird species infected experimentally with low-pathogenicity avian influenza viruses that originated from wild birds. Avian Pathol 40(2):119–124
Stallknecht DE, Brown JD (2008) Ecology of avian influenza in wild birds. In: Swayne D (ed) Blackwell, Iowa
Stallknecht DE, Brown JD (2009) Tenacity of avian influenza viruses. Rev Sci Tech 28(1):59–67
Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E (2009) Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83(9):4642–4651
Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E (2010) Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84(14):6955–6965
Day JM, Ballard LL, Duke MV, Scheffler BE, Zsak L (2010) Metagenomic analysis of the turkey gut RNA virus community. Virol J 7:313
Shan T, Li L, Simmonds P, Wang C, Moeser A, Delwart E (2011) The fecal virome of pigs on a high-density farm. J Virol 85(22):11697–11708
Phan TG, Vo NP, Ak B, Pankovics P, Reuter G, Li OT, Wang C, Deng X, Poon LL, Delwart E (2013) The viruses of wild pigeon droppings. PLoS ONE 8(9):e72787
Fawaz M, Vijayakumar P, Mishra A, Gandhale PN, Dutta R, Kamble NM, Sudhakar SB, Roychoudhary P, Kumar H, Kulkarni DD, Raut AA (2016) Duck gut viral metagenome analysis captures snapshot of viral diversity. Gut Pathog 8:30
Morgan RW, Cantello JL, McDermott CH (1990) Transfection of chicken embryo fibroblasts with Marek’s disease virus DNA. Avian Dis 34(2):345–351
Wang H, Song S, Zeng J, Zhou G, Yang D, Liang T, Yu L (2014) Single amino acid substitution of VP1 N17D or VP2 H145Y confers acid-resistant phenotype of type Asia1 foot-and-mouth disease virus. Virol Sin 29(2):103–111
Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J (2001) A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci USA 98(20):11609–11614
Cheng WX, Li JS, Huang CP, Yao DP, Liu N, Cui SX, Jin Y, Duan ZJ (2010) Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS ONE 5(10):e13583
Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, Kapczynski DR (2017) Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509:159–166
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform 9:386
Richard M, Fouchier RA (2016) Influenza A virus transmission via respiratory aerosols or droplets as it relates to pandemic potential. FEMS Microbiol Rev 40(1):68–85
Talker SC, Stadler M, Koinig HC, Mair KH, Rodriguez-Gomez IM, Graage R, Zell R, Durrwald R, Starick E, Harder T, Weissenbock H, Lamp B, Hammer SE, Ladinig A, Saalmuller A, Gerner W (2016) Influenza A virus infection in pigs attracts multifunctional and cross-reactive T cells to the lung. J Virol 90(20):9364–9382
Li Y, Huang B, Ma X, Wu J, Li F, Ai W, Song M, Yang H (2009) Molecular characterization of the genome of duck enteritis virus. Virology 391(2):151–161
Dhama K, Kumar N, Saminathan M, Tiwari R, Karthik K, Kumar MA, Palanivelu M, Shabbir MZ, Malik YS, Singh RK (2017) Duck virus enteritis (duck plague)—a comprehensive update. Vet Q 37(1):57–80
Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F (2008) Viral diversity and dynamics in an infant gut. Res Microbiol 159(5):367–373
Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F (2003) Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185(20):6220–6223
Cann AJ, Fandrich SE, Heaphy S (2005) Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30(2):151–156
Li L, Shan T, Wang C, Cote C, Kolman J, Onions D, Gulland FM, Delwart E (2011) The fecal viral flora of California sea lions. J Virol 85(19):9909–9917
Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, Shaukat S, Masroor MA, Wilson ML, Ndjango JB, Peeters M, Gross-Camp ND, Muller MN, Hahn BH, Wolfe ND, Triki H, Bartkus J, Zaidi SZ, Delwart E (2010) Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84(4):1674–1682
Manteufel J, Truyen U (2008) Animal bocaviruses: a brief review. Intervirology 51(5):328–334
Cagle C, To TL, Nguyen T, Wasilenko J, Adams SC, Cardona CJ, Spackman E, Suarez DL, Pantin-Jackwood MJ (2011) Pekin and Muscovy ducks respond differently to vaccination with a H5N1 highly pathogenic avian influenza (HPAI) commercial inactivated vaccine. Vaccine 29(38):6549–6557
Cagle C, Wasilenko J, Adams SC, Cardona CJ, To TL, Nguyen T, Spackman E, Suarez DL, Smith D, Shepherd E, Roth J, Pantin-Jackwood MJ (2012) Differences in pathogenicity, response to vaccination, and innate immune responses in different types of ducks infected with a virulent H5N1 highly pathogenic avian influenza virus from Vietnam. Avian Dis 56(3):479–487
Acknowledgements
This work was supported by the National Key R&D Program of China (2016YFD0500800) and the Chinese Academy of Agricultural Sciences Fundamental Scientific Research Funds (Y2016PT41).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Ethical Approval
This study was approved by Harbin Veterinary Research Institute and performed in accordance with ethics guidelines and approved protocols. The animal Ethics Committee approval number is Heilongjiang-SYXK-2006-032.
Rights and permissions
About this article
Cite this article
Zhao, L., Niu, Y., Lu, T. et al. Metagenomic Analysis of the Jinding Duck Fecal Virome. Curr Microbiol 75, 658–665 (2018). https://doi.org/10.1007/s00284-018-1430-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1430-3