Abstract
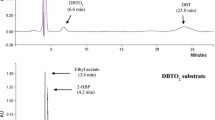
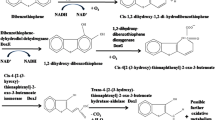
Microorganisms can metabolize or transform a range of known chemical compounds present in fossil fuels by naturally having highly specific metabolic activities. In this context, the microbial desulfurization of fuels is an attractive and alternative process to the conventional hydrodesulfurization (HDS) process, since the thiophenic sulfur containing compounds such as dibenzothiophene (DBT) and benzothiophene (BT) cannot be removed by HDS. A DBT desulfurizing mesophilic bacterium, identified on the basis of 16S rRNA gene sequence as Gordonia sp. HS126-4N (source: periphery soil of a coal heap) has been evaluated for its biodesulfurization traits and potential to desulfurize the thiophenic compounds. The HPLC and LC/MS analyses of the metabolites produced from DBT desulfurization and PCR-based nucleotide sequence confirmation of the key desulfurizing genes (dszA/dszB/dszC) proved that HS126-4N could convert DBT to 2-hydroxybiphenyl (2-HBP) via the 4S pathway. The isolate could convert 0.2 mM of DBT to 2-HBP within 48 h and was reasonably tolerant against the inhibitory effect of 2-HBP (retained 70% of growth at 0.5 mM 2-HBP). The isolated biocatalyst desulfurized/degraded 100% of 0.2 mM of 4-methyl DBT, 2,8-dimethyl DBT, BT and 3-methyl BT within 108 h. The capabilities to survive and desulfurize a broad range of thiophenic sulfur containing substrates as well as less inhibition by the 2-HBP suggest that HS126-4N could be a potential candidate for improved biodesulfurization/organic sulfur removal from fossil fuels.



Similar content being viewed by others
References
Abin-Fuentes A, Mohamed ME, Wang DIC, Prather KLJ (2013) Exploring the mechanism of biocatalyst inhibition in microbial desulfurization. Appl Environ Microbiol 79:7807–7817
Aggarwal S, Karimi IA, Ivan GR (2013) In silico modeling and evaluation of Gordonia alkanivorans for biodesulfurization. Mol Biosyst 9:2530–2540
Akhtar N, Ghauri MA, Akhtar K (2016) Dibenzothiophene desulfurization capability and evolutionary divergence of newly isolated bacteria. Arch Microbiol 198:509–519
Akhtar N, Ghauri MA, Anwar MA, Akhtar K (2009) Analysis of the dibenzothiophene metabolic pathway in a newly isolated Rhodococcus spp. FEMS Microbiol Lett 301:95–102
Akhtar N, Ghauri MA, Anwar MA, Heaphy S (2015) Phylogenetic characterization and novelty of organic sulfur metabolizing genes of Rhodococcus spp. (Eu-32). Biotechnol Lett 37:837–847
Alves L, Salgueiro R, Rodrigues C, Mesquita E, Matos J, Girio FM (2005) Desulfurization of dibenzothiophene, benzothiophene, and other thiophene analogs by a newly isolated bacterium, Gordonia alkanivorans strain 1B. Appl Biochem Biotechnol 120:199–208
Bahuguna A, Lily MK, Munjal A, Singh RN, Dangwal K (2011) Desulfurization of dibenzothiophene (DBT) by a novel strain Lysinibacillus sphaericus DMT-7 isolated from diesel contaminated soil. J Environ Sci 23:975–982
Bhatia S, Sharma DK (2010) Biodesulfurization of dibenzothiophene, its alkylated derivatives and crude oil by a newly isolated strain Pantoea agglomerans D23W3. Biochem Eng J 50:104–109
Carvajal P, Dinamarca MA, Baeza P, Camú E, Ojeda J (2017) Removal of sulfur-containing organic molecules adsorbed on inorganic supports by Rhodococcus Rhodochrous spp. Biotechnol Lett 39:241–245
Chen H, Zhang WJ, Cai YB, Zhang Y, Li W (2008) Elucidation of 2-hydroxybiphenyl effect on dibenzothiophene desulfurization by Microbacterium sp. strain ZD-M2. Bioresource Technol 99:6928–69233
Drzyzga O (2012) The strengths and weaknesses of Gordonia: a review of an emerging genus with increasing biotechnological potential. Crit Rev Microbiol 38:300–316
Feng S, Yang H, Zhan X, Wang W (2016) Enhancement of dibenzothiophene biodesulfurization by weakening the feedback inhibitions effects based on a systematic understanding of the biodesulfurization mechanism by Gordonia sp. through the potential “4S” pathway. RSC Adv 6:82872–82881
Gallagher JR, Olson ES, Stanley DC (1993) Microbial desulfurization of dibenzothiophene: a sulfur-specific pathway. FEMS Microbiol Lett 107:31–35
Gilbert SC, Morton J, Buchanan S, Oldfield C, McRoberts A (1998) Isolation of a unique benzothiophene-desulphurizing bacterium, Gordonia sp. strain 213E (NCIMB 40816), and characterization of the desulphurization pathway. Microbiology 144:2545–2553
Gunam IBW, Yaku Y, Hirano M, Yamamura K, Tomita F, Sone T, Asano K (2006) Biodesulfurization of alkylated forms of dibenzothiophene and benzothiophene by Sphingomonas subarctica T7b. J Biosci Bioeng 101:322–327
Ishii Y, Kozaki S, Furuya T, Kino K, Kirimura K (2005) Thermophilic biodesulfurization of various heterocyclic sulfur compounds and crude straight-run light gas oil fraction by a newly isolated strain Mycobacterium phlei WU-0103. Curr Microbiol 50:63–70
Kilbane JJ II (2017) Biodesulfurization: how to make it work? Arabian J Sci Eng 42:1–9
Kilbane JJ, Robbins J (2007) Characterization of the dszABC genes of Gordonia amicalis F.5.25.8 and identification of conserved protein and DNA sequences. Appl Microbiol Biotechnol 75:843–851
Konishi J, Onaka T, Ishii Y, Suzuki M (2000) Demonstration of the carbon-sulfur bond targeted desulfurization of benzothiophene by thermophilic Paenibacillus sp. strain A11–2 capable of desulfurizing dibenzothiophene. FEMS Microbiol Lett 187:151–154
Li F, Xu P, Feng J, Meng L, Zheng Y, Luo L, Ma C (2005) Microbial desulfurization of gasoline in a Mycobacterium goodii X7B immobilized-cell system. Appl Environ Microbiol 71:276–281
Li W, Zhang Y, Wang MD, Shi Y (2005) Biodesulfurization of dibenzothiophene and other organic sulfur compounds by a newly isolated Microbacterium strain ZD-M2. FEMS Microbiol Lett 247:45–50
Mishra S, Pradhan N, Panda S, Akcil A (2016) Biodegradation of dibenzothiophene and its application in the production of clean coal. Fuel Process Technol 152:325–342
Mohamed ME-S, Al-Yacoub ZH, Vedakumar JV (2015) Biocatalytic desulfurization of thiophenic compounds and crude oil by newly isolated bacteria. Front Microbiol 6:1–12
Mohebali G, Ball AS (2008) Biocatalytic desulfurization (BDS) of petrodiesel fuels. Microbiol 154:2169–2183
Piddington CS, Kovacevich BR, Rambosek J (1995) Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp strain IGTS8. Appl Environ Microbiol 61:468–475
Soleimani M, Bassi A, Margaritis A (2007) Biodesulfurization of refractory organic sulfur compounds in fossil fuels. Biotechnol Adv 25:570–596
Tanaka Y, Matsui T, Konishi J, Maruhashi K, Kurane R (2002) Biodesulfurization of benzothiophene and dibenzothiophene by a newly isolated Rhodococcus strain. Appl Microbiol Biotechnol 59:325–328
Tanaka Y, Onaka T, Matsui T, Maruhashi K, Kurane R (2001) Desulfurization of benzothiophene by the Gram-negative bacterium, Sinorhizobium sp. KT55. Curr Microbiol 43:187–191
Wang W, Ma T, Lian K, Zhang Y, Tian H, Ji K, Li G (2013) Genetic analysis of benzothiophene biodesulfurization pathway of Gordonia terrae strain C-6. PLoS ONE 8:e84386
Acknowledgements
This research work was supported by International Foundation for Science (IFS), Stockholm, Sweden through a Grant to Dr. Nasrin Akhtar (Agreement No. F/5379-1) and co-supported by the Committee on Scientific and Technological Cooperation (COMSTECH), Pakistan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akhtar, N., Akhtar, K. & Ghauri, M.A. Biodesulfurization of Thiophenic Compounds by a 2-Hydroxybiphenyl-Resistant Gordonia sp. HS126-4N Carrying dszABC Genes. Curr Microbiol 75, 597–603 (2018). https://doi.org/10.1007/s00284-017-1422-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1422-8