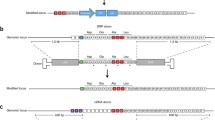
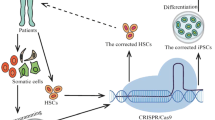
Abstract
Gene therapy represents a significant potential to revolutionize the field of hematology with applications in correcting genetic mutations, generating cell lines and animal models, and improving the feasibility and efficacy of cancer immunotherapy. Compared to different genetic engineering tools, clustered regularly interspaced short palindromic repeats (CRISPR) CRISPR-associated protein 9 (Cas9) emerged as an effective and versatile genetic editor with the ability to precisely modify the genome. The applications of genetic engineering in various hematological disorders have shown encouraging results. Monogenic hematological disorders can conceivably be corrected with single gene modification. Through the use of CRISPR-CAS9, restoration of functional red blood cells and hemostasis factors were successfully attained in sickle cell anemia, beta-thalassemia, and hemophilia disorders. Our understanding of hemato-oncology has been advanced via CRIPSR-CAS9 technology. CRISPR-CAS9 aided to build a platform of mutated genes responsible for cell survival and proliferation in leukemia. Therapeutic application of CRISPR-CAS9 when combined with chimeric antigen receptor (CAR) T cell therapy in multiple myeloma and acute lymphoblastic leukemia was feasible with attenuation of CAR T cell therapy pitfalls. Our review outlines the latest literature on the utilization of CRISPR-Cas9 in the treatment of beta-hemoglobinopathies and hemophilia disorders. We present the strategies that were employed and the findings of preclinical and clinical trials. Also, the review will discuss gene engineering in the field of hemato-oncology as a proper tool to facilitate and overcome the drawbacks of chimeric antigen receptor T cell therapy (CAR-T).



Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Abbreviations
- DNA :
-
deoxyribonucleic acid
- ZFN :
-
zinc finger nuclease
- TALEN :
-
transcription activator-like effector nuclease
- CRISPR :
-
clustered regulatory interspaced short palindromic repeats
- Cas9 :
-
CRISPR-associated protein 9
- DSB :
-
DNA double-strand break
- NHEJ :
-
non-homologous end joining
- HDR :
-
homology-directed repair
- INDELs :
-
insertion and deletion mutations
- RNA :
-
ribonucleic acid
- SCD :
-
sickle cell disease
- HSCT :
-
hematopoietic stem cell transplantation
- Cr-RNA :
-
CRISPR RNA
- Tracr-RNA :
-
transactivating crRNA
- PAM :
-
protospacer adjacent motif
- sgRNA :
-
single-strand small-guide RNA
- CAR :
-
chimeric antigen receptor
- dCas9 :
-
deactivated Cas9
- RT :
-
reverse transcriptase
- pegRNA :
-
prime-editing single guide RNA
- SCA :
-
sickle cell anemia
- TM :
-
thalassemia major
- HbS :
-
hemoglobin S
- HbF :
-
fetal hemoglobin
- HbA :
-
adult hemoglobin
- HPFH :
-
hereditary persistence of fetal hemoglobin
- IPSCs :
-
induced pluripotent stem cells
- UTR :
-
untranslated region
- FDA :
-
Food and Drug Administration
- HLA :
-
human leukocyte antigens
- TRAC :
-
T-cell alpha receptor
- PD-1 :
-
programmed cell death protein 1
- GvH :
-
graft-versus-host
- TCR-B :
-
T cell receptor beta
- ASXL1 :
-
additional sex combs-like 1
- AML :
-
acute myeloid leukemia
- CML :
-
chronic myeloid leukemia
- PRC2 :
-
polycomb repressive complex 2
- VRPEB1 :
-
V-set pre-B-cell surrogate light chain 1
- MM :
-
multiple myeloma
- BDDF8 :
-
B domain-deleted factor 8
- EF1α :
-
human elongation factor 1 alpha
- ssODN :
-
single-stranded-oligodeoxynucleotide
References
Jiang S, Shen QW (2019) Principles of gene editing techniques and applications in animal husbandry. 3. Biotech 9(1):1–9
Thomas KR, Capecchi MR (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 51(3):503–512
González-Romero E, Martínez-Valiente C, García-Ruiz C, Vázquez-Manrique RP, Cervera J, Sanjuan-Pla A (2019) CRISPR to fix bad blood: a new tool in basic and clinical hematology. Haematologica. 104(5):881
Ahmad HI, Ahmad MJ, Asif AR, Adnan M, Iqbal MK, Mehmood K, Muhammad SA, Bhuiyan AA, Elokil A, Du X, Zhao C (2018) A review of CRISPR-based genome editing: survival, evolution and challenges. Curr Issues Mol Biol 28(1):47–68
Chen Y, Wen R, Yang Z, Chen Z (2022) Genome editing using CRISPR/Cas9 to treat hereditary hematological disorders. Gene Ther 29(5):207–216
Asmamaw M, Zawdie B (2021) Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biol Targets Ther 15:353
Jacinto FV, Link W, Ferreira BI (2020) CRISPR/Cas9-mediated genome editing: from basic research to translational medicine. J Cell Mol Med 24(7):3766–3778
Konstantakos V, Nentidis A, Krithara A, Paliouras G (2022) CRISPR–Cas9 gRNA efficiency prediction: an overview of predictive tools and the role of deep learning. Nucleic Acids Res 50(7):3616–3637
Tahir T, Ali Q, Rashid MS, Malik A (2020) The journey of CRISPR-Cas9 from bacterial defense mechanism to a gene editing tool in both animals and plants. Biol Clin Sci Res J 2020(1)
Westermann L, Neubauer B, Köttgen M (2021) Nobel Prize 2020 in Chemistry honors CRISPR: a tool for rewriting the code of life. Pflugers Arch - Eur J Physiol 473(1):1–2
Porteus MH (2017) Genome editing for the β-hemoglobinopathies. In: Malik P, Tisdale J (eds) Gene and cell therapies for beta-globinopathies. Advances in experimental medicine and biology, vol 1013. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-7299-9_8
Kunz JB, Kulozik AE (2020) Gene therapy of the hemoglobinopathies. HemaSphere 4:5
Frati G, Miccio A (2021) Genome editing for β-hemoglobinopathies: advances and challenges. J Clin Med 10(3):482
Sochacka-Ćwikła A, Mączyński M, Regiec A (2021) FDA-approved drugs for hematological malignancies—the last decade review. Cancers 14(1):87
Stewart RL, Koo SC, Furtado LV (2018) Principles of molecular biology and oncogenesis. In Precision molecular pathology of neoplastic pediatric diseases. Springer, Cham, pp 3–8
Reschke M, Seitzer N, Clohessy J, Pandolfi P (2014) Hematological malignancies and premalignant conditions. In: Parsyan A (ed) Translation and its regulation in cancer biology and medicine. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-9078-9_23
Dakubo GD (2019) Hematologic malignancy biomarkers in proximal fluids. In: Cancer biomarkers in body fluids. Springer, Cham. https://doi.org/10.1007/978-3-030-24725-6_12
Deshantri AK, Moreira AV, Ecker V, Mandhane SN, Schiffelers RM, Buchner M, Fens MH (2018) Nanomedicines for the treatment of hematological malignancies. J Control Release 287:194–215
Berntorp E, Fischer K, Hart DP, Mancuso ME, Stephensen D, Shapiro AD, Blanchette V (2021) Haemophilia. Nat Rev Dis Primers 7(1):1–9
Weyand AC, Pipe SW (2019) New therapies for hemophilia. Blood J American Soc Hematol 133(5):389–398
Park CY, Lee DR, Sung JJ, Kim DW (2016) Genome-editing technologies for gene correction of hemophilia. Hum Genet 135(9):977–981
Lone BA, Karna SKL, Ahmad F, Shahi N, Pokharel YR (2018) CRISPR/CAS9 System: a bacterial tailor for genomic engineering. Genet Res Int 2018:1–17
Hryhorowicz M, Lipiński D, Zeyland J, Słomski R (2017) CRISPR/Cas9 immune system as a tool for genome engineering. Arch Immunol Ther Exp 65(3):233–240
Jiang F, Doudna JA (2017) CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys 46(1):505–529
Wang SW, Gao C, Zheng YM et al (2022) Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer 21:57. https://doi.org/10.1186/s12943-022-01518-8
Gupta D, Bhattacharjee O, Mandal D, Sen MK, Dey D, Dasgupta A, Kazi TA, Gupta R, Sinharoy S, Acharya K, Chattopadhyay D (2019) CRISPR-Cas9 system: a new-fangled dawn in gene editing. Life Sci 232:116636
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213):1258096
Liu C, Zhang L, Liu H, Cheng K (2017 Nov) Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release 28(266):17–26
Janik E, Niemcewicz M, Ceremuga M, Krzowski L, Saluk-Bijak J, Bijak M (2020) Various aspects of a gene editing system—crispr–cas9. Int J Mol Sci 21(24):9604
Zhang B (2021) CRISPR/Cas gene therapy. J Cell Physiol 236(4):2459–2481
Zhang H, Qin C, An C, Zheng X, Wen S, Chen W, Liu X, Lv Z, Yang P, Xu W, Gao W (2021) Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer 20(1):1–22
Ghaemi A, Bagheri E, Abnous K, Taghdisi SM, Ramezani M, Alibolandi M (2021) CRISPR-cas9 genome editing delivery systems for targeted cancer therapy. Life Sci 267:118969
Hendriks D, Clevers H, Artegiani B (2020) CRISPR-Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 27(5):705–731
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533(7603):420–424
Memi F, Ntokou A, Papangeli I. CRISPR/Cas9 gene-editing: research technologies, clinical applications and ethical considerations. Seminars in perinatology 2018 (42 8, 487-500). WB Saunders
Kantor A, McClements ME, MacLaren RE (2020) CRISPR-Cas9 DNA base-editing and prime-editing. Int J Mol Sci 21(17):6240
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576(7785):149–157
Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38(7):824–844
Sundd P, Gladwin MT, Novelli EM (2019) Pathophysiology of sickle cell disease. Annu Rev Pathol 14:263
Williams TN, Thein SL (2018) Sickle cell anemia and its phenotypes. Annu Rev Genomics Hum Genet 19:113–147
Tisdale JF, Thein SL, Eaton WA (2020) Treating sickle cell anemia. Science 367(6483):1198–1199
Needs T, Gonzalez-Mosquera LF, Lynch DT (2022) Beta thalassemia. StatPearls [Internet]. StatPearls Publishing, Treasure Island (FL)
Constantinou C, Spella M, Chondrou V, Patrinos GP, Papachatzopoulou A, Sgourou A (2019) The multi-faceted functioning portrait of LRF/ZBTB7A. Hum Gen 13(1):1–4
Demirci S, Zeng J, Wu Y, Uchida N, Shen AH, Pellin D, Gamer J, Yapundich M, Drysdale C, Bonanno J, Bonifacino AC (2020) BCL11A enhancer–edited hematopoietic stem cells persist in rhesus monkeys without toxicity. J Clin Invest 130(12):6677–6687
Bauer DE, Orkin SH (2015) Hemoglobin switching’s surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr Opin Genet Dev 33:62–70
Jensen TI, Axelgaard E, Bak RO (2019) Therapeutic gene editing in haematological disorders with CRISPR/Cas9. Br J Haematol 185(5):821–835
Quintana-Bustamante O, Fañanas-Baquero S, Dessy-Rodriguez M, Ojeda-Pérez I, Segovia JC (2022) Gene editing for inherited red blood cell diseases. Front Physiol 28(13):848261
Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S (2015) BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527(7577):192–197
Zeng J, Wu Y, Ren C, Bonanno J, Shen AH, Shea D, Gehrke JM, Clement K, Luk K, Yao Q, Kim R (2020) Therapeutic base editing of human hematopoietic stem cells. Nat Med 26(4):535–541
Wu Y, Zeng J, Roscoe BP, Liu P, Yao Q, Lazzarotto CR, Clement K, Cole MA, Luk K, Baricordi C, Shen AH (2019) Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 25(5):776–783
Weber L, Frati G, Felix T, Hardouin G, Casini A, Wollenschlaeger C, Meneghini V, Masson C, De Cian A, Chalumeau A, Mavilio F (2020) Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci Adv 6(7):eaay9392
Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ, Zhu C (2016) Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351(6270):285–289
Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, Foell J, de la Fuente J, Grupp S, Handgretinger R, Ho TW (2021) CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med 384(3):252–260
Fu B, Liao J, Chen S, Li W, Wang Q, Hu J, Yang F, Hsiao S, Jiang Y, Wang L, Chen F (2022) CRISPR–Cas9-mediated gene editing of the BCL11A enhancer for pediatric β0/β0 transfusion-dependent β-thalassemia. Nat Med 28(8):1573–1580
Esrick EB, Lehmann LE, Biffi A, Achebe M, Brendel C, Ciuculescu MF, Daley H, MacKinnon B, Morris E, Federico A, Abriss D (2021) Post-transcriptional genetic silencing of BCL11A to treat sickle cell disease. N Engl J Med 384(3):205–215
Cao A, Galanello R (2010) Beta-thalassemia. Genet Med 12(2):61–76
Thein SL (2017) Genetic basis and genetic modifiers of β-thalassemia and sickle cell disease. Gene and Cell Therapies for Beta-Globinopathies 27–57
Song B, Fan Y, He W, Zhu D, Niu X, Wang D, Ou Z, Luo M, Sun X (2015) Improved hematopoietic differentiation efficiency of gene-corrected beta-thalassemia induced pluripotent stem cells by CRISPR/Cas9 system. Stem Cells Dev 24(9):1053–1065
Niu X, He W, Song B, Ou Z, Fan D, Chen Y, Fan Y, Sun X (2016) Combining single strand oligodeoxynucleotides and CRISPR/Cas9 to correct gene mutations in β-thalassemia-induced pluripotent stem cells. J Biol Chem 291(32):16576–16585
Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW (2014) Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 24(9):1526–1533
Xu P, Tong Y, Liu XZ, Wang TT, Cheng L, Wang BY, Lv X, Huang Y, Liu DP (2015) Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C> T) mutation in β-thalassemia-derived iPSCs. Sci Rep 5(1):1–2
Dever DP et al (2016) CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539(7629):384–389
Wilkinson AC, Dever DP, Baik R, Camarena J, Hsu I, Charlesworth CT, Morita C, Nakauchi H, Porteus MH (2021) Cas9-AAV6 gene correction of beta-globin in autologous HSCs improves sickle cell disease erythropoiesis in mice. Nat Commun 12(1):1–9
Lattanzi A, Camarena J, Lahiri P, Segal H, Srifa W, Vakulskas CA, Frock RL, Kenrick J, Lee C, Talbott N, Skowronski J (2021) Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci Transl Med 13(598):eabf2444
Cai L, Bai H, Mahairaki V, Gao Y, He C, Wen Y, Jin YC, Wang Y, Pan RL, Qasba A, Ye Z (2018) A universal approach to correct various HBB gene mutations in human stem cells for gene therapy of beta-thalassemia and sickle cell disease. Stem Cells Transl Med 7(1):87–97
Li H, Yang Y, Hong W et al (2020) Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther 5:1. https://doi.org/10.1038/s41392-019-0089-y
Iyer DN, Schimmer AD, Chang H (2023) Applying CRISPR-Cas9 screens to dissect hematological malignancies. Blood Adv 7(10):2252–2270
Sterner RC, Sterner RM (2021) CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J 11(4):1
Mohanty R, Chowdhury CR, Arega S, Sen P, Ganguly P, Ganguly N (2019) CAR T cell therapy: a new era for cancer treatment. Oncol Rep 42(6):2183–2195
Han D, Xu Z, Zhuang Y, Ye Z, Qian Q (2021) Current progress in CAR-T cell therapy for hematological malignancies. J Cancer 12(2):326
Holstein SA, Lunning MA (2020) CAR T-cell therapy in hematologic malignancies: a voyage in progress. Clin Pharmacol Ther 107(1):112–122
Dimitri A, Herbst F, Fraietta JA (2022) Engineering the next-generation of CAR T-cells with CRISPR-Cas9 gene editing. Mol Cancer 21(1):1–3
Razeghian E, Nasution MK, Rahman HS, Gardanova ZR, Abdelbasset WK, Aravindhan S, Bokov DO, Suksatan W, Nakhaei P, Shariatzadeh S, Marofi F (2021) A deep insight into CRISPR/Cas9 application in CAR-T cell-based tumor immunotherapies. Stem Cell Res Ther 12(1):1–7
Khan A, Sarkar E (2022) Review CRISPR/Cas9 encouraged CAR-T cell immunotherapy reporting efficient and safe clinical results towards cancer. Cancer Treat Res Commun 27:100641
Wang L, Chen Y, Liu X, Li Z, Dai X (2022) The application of CRISPR/Cas9 technology for cancer immunotherapy: current status and problems. Front Oncol 17(11):5853
Sterner RM, Sakemura R, Cox MJ, Yang N, Khadka RH, Forsman CL, Hansen MJ, Jin F, Ayasoufi K, Hefazi M, Schick KJ (2019) GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood J American Soc Hematol 133(7):697–709
Tang N, Cheng C, Zhang X, Qiao M, Li N, Mu W, Wei XF, Han W, Wang H (2020) TGF-β inhibition via CRISPR promotes the long-term efficacy of CAR T cells against solid tumors. JCI insight 5(4)
Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, Odak A, Gönen M, Sadelain M (2017) Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543(7643):113–117
Georgiadis C, Preece R, Nickolay L, Etuk A, Petrova A, Ladon D, Danyi A, Humphryes-Kirilov N, Ajetunmobi A, Kim D, Kim JS (2018) Long terminal repeat CRISPR-CAR-coupled “universal” T cells mediate potent anti-leukemic effects. Mol Ther 26(5):1215–1227
Rupp LJ, Schumann K, Roybal KT, Gate RE, Ye CJ, Lim WA, Marson A (2017) CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci Rep 7(1):1
Stenger D, Stief TA, Kaeuferle T, Willier S, Rataj F, Schober K, Vick B, Lotfi R, Wagner B, Grünewald TG, Kobold S (2020) Endogenous TCR promotes in vivo persistence of CD19-CAR-T cells compared to a CRISPR/Cas9-mediated TCR knockout CAR. Blood 136(12):1407–1418
Stadtmauer EA, Cohen AD, Weber K, Lacey SF, Gonzalez VE, Melenhorst JJ, June CH (2019) First-in-human assessment of feasibility and safety of multiplexed genetic engineering of autologous T cells expressing NY-ESO-1 TCR and CRISPR/Cas9 gene edited to eliminate endogenous TCR and PD-1 (NYCE T cells) in advanced multiple myeloma (MM) and sarcoma. Blood 134:49
Wei G, Zhang Y, Zhao H, Wang Y, Liu Y, Liang B, Wang X, Xu H, Cui J, Wu W, Zhao K (2021) CD19/CD22 dual-targeted CAR T-cell therapy for relapsed/refractory aggressive B-cell lymphoma: a safety and efficacy study. Cancer Immunol Res 9(9):1061–1070
Asada S, Fujino T, Goyama S, Kitamura T (2019) The role of ASXL1 in hematopoiesis and myeloid malignancies. Cell Mol Life Sci 76(13):2511–2523
Valletta S, Dolatshad H, Bartenstein M, Yip BH, Bello E, Gordon S, Yu Y, Shaw J, Roy S, Scifo L, Schuh A (2015) ASXL1 mutation correction by CRISPR/Cas9 restores gene function in leukemia cells and increases survival in mouse xenografts. Oncotarget 6(42):44061
Felce SL, Anderson AP, Maguire S, Gascoyne DM, Armstrong RN, Wong KK, Li D, Banham AH (2020) CRISPR/Cas9-mediated Foxp1 silencing restores immune surveillance in an immunocompetent A20 lymphoma model. Front Oncol 3(10):448
Schlager S, Salomon C, Olt S, Albrecht C, Ebert A, Bergner O, Wachter J, Trapani F, Gerlach D, Voss T, Traunbauer A (2020) Inducible knock-out of BCL6 in lymphoma cells results in tumor stasis. Oncotarget 11(9):875
Khaled M, Moustafa AS, El-Khazragy N, Ahmed MI, Abd Elkhalek MA, El-Salahy EM (2021) CRISPR/Cas9 mediated knock-out of VPREB1 gene induces a cytotoxic effect in myeloma cells. PLoS One 16(1):e0245349
Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, Gozdecka M (2016) A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep 17(4):1193–1205
Mercier FE, Shi J, Sykes DB, Oki T, Jankovic M, Man CH, Kfoury YS, Miller E, He S, Zhu A, Vasic R (2022) In vivo genome-wide CRISPR screening in murine acute myeloid leukemia uncovers microenvironmental dependencies. Blood Adv 6(17):5072–5084
Ohmori T (2020) Advances in gene therapy for hemophilia: basis, current status, and future perspectives. Int J Hematol 111(1):31–41
Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS (2015) Functional correction of large factor VIII gene chromosomal inversions in hemophilia A patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell 17(2):213–220
Dinh LV, Fine E, Ameri A, Luu BW, Kumar S, Diego VP, Curran JE, Peralta JM, Mead H, Escobar MA, Williams-Blangero S (2020) Specific correction of the intron-22 inverted factor VIII gene in autologous blood outgrowth endothelial cells from patients with severe hemophilia A. Blood 5(136):30–31
Luo S, Li Z, Dai X, Zhang R, Liang Z, Li W, Zeng M, Su J, Wang J, Liang X, Wu Y (2021) CRISPR/Cas9-mediated in vivo genetic correction in a mouse model of hemophilia A. Front Cell Dev Biol 16(9):672564
Tantawy AA (2010) Molecular genetics of hemophilia A: Clinical perspectives. Egypt J Med Hum Genet 11(2)
Everett LA, Cleuren AC, Khoriaty RN, Ginsburg D (2014) Murine coagulation factor VIII is synthesized in endothelial cells. Blood J American Soc Hematol 123(24):3697–3705
Shahani T, Covens K, Lavend'Homme R, Jazouli N, Sokal E, Peerlinck K, Jacquemin M (2014) Human liver sinusoidal endothelial cells but not hepatocytes contain factor VIII. J Thromb Haemost 12(1):36–42
Zhang JP, Cheng XX, Zhao M, Li GH, Xu J, Zhang F, Yin MD, Meng FY, Dai XY, Fu YW, Yang ZX (2019) Curing hemophilia A by NHEJ-mediated ectopic F8 insertion in the mouse. Genome Biol 20(1):1–7
Chen H, Shi M, Gilam A, Zheng Q, Zhang Y, Afrikanova I, Li J, Gluzman Z, Jiang R, Kong LJ, Chen-Tsai RY (2019) Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Sci Rep 9(1):1–5
Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, Wang L, Zeng L, Shao Y, Chen Y, Ma N (2016) CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med 8(5):477–488
Sung JJ, Park CY, Leem JW et al (2019) Restoration of FVIII expression by targeted gene insertion in the FVIII locus in hemophilia A patient-derived iPSCs. Exp Mol Med 51:1–9
Hu Z, Zhou M, Wu Y, Li Z, Liu X, Wu L, Liang D (2019) ssODN-mediated in-frame deletion with CRISPR/Cas9 restores FVIII function in hemophilia A-patient-derived iPSCs and ECs. Mol Ther Nucleic Acids 6(17):198–209
Rezaie AR, Giri H (2020) Anticoagulant and signaling functions of antithrombin. J Thromb Haemost 18(12):3142–3153
Han JP, Kim M, Choi BS, Lee JH, Lee GS, Jeong M, Lee Y, Kim EA, Oh HK, Go N, Lee H (2022) In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci Adv 8(3):eabj6901
Hu YF, Fang YH, Lai YR, Feng XQ, Xu SQ (2022) Application of gene therapy in hemophilia. Curr Med Sci 19:1–7
Sun J, Wang J, Zheng D, Hu X (2020) Advances in therapeutic application of CRISPR-Cas9. Brief Funct Genom 19(3):164–174
Tsai SQ, Joung JK (2016) Defining and improving the genome-wide specificities of CRISPR–Cas9 nucleases. Nat Rev Genet 17(5):300–312
Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM (2016) A multifunctional AAV–CRISPR–Cas9 and its host response. Nat Methods 13(10):868–874
Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, Behlke MA (2019) Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med 25(2):249–254
Ferdosi SR, Ewaisha R, Moghadam F, Krishna S, Park JG, Ebrahimkhani MR, Kiani S, Anderson KS (2019) Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun 10(1):1
Pavani G, Fabiano A, Laurent M, Amor F, Cantelli E, Chalumeau A, Maule G, Tachtsidi A, Concordet JP, Cereseto A, Mavilio F (2021) Correction of β-thalassemia by CRISPR/Cas9 editing of the α-globin locus in human hematopoietic stem cells. Blood Adv 5(5):1137–1153
Li L, Yi H, Liu Z, Long P, Pan T, Huang Y, Li Y, Li Q, Ma Y (2022) Genetic correction of concurrent α-and β-thalassemia patient-derived pluripotent stem cells by the CRISPR-Cas9 technology. Stem Cell Res Ther 13(1):102
Cosenza LC, Gasparello J, Romanini N, Zurlo M, Zuccato C, Gambari R, Finotti A (2021) Efficient CRISPR-Cas9-based genome editing of β-globin gene on erythroid cells from homozygous β039-thalassemia patients. Mol Ther Methods Clin Dev 11(21):507–523
Uchida N, Drysdale CM, Nassehi T, Gamer J, Yapundich M, DiNicola J, Shibata Y, Hinds M, Gudmundsdottir B, Haro-Mora JJ, Demirci S (2021) Cas9 protein delivery non-integrating lentiviral vectors for gene correction in sickle cell disease. Mol Ther Methods Clin Dev 11(21):121–132
Li C, Wang H, Georgakopoulou A, Gil S, Yannaki E, Lieber A (2021) In vivo HSC gene therapy using a bi-modular HDAd5/35++ vector cures sickle cell disease in a mouse model. Mol Ther 29(2):822–837
Patel RP, Ghilardi G, Porazzi P, Yang S, Qian D, Pajarillo R, Wang M, Zhang Y, Schuster SJ, Barta SK, Nimmagadda A (2022) Clinical development of Senza5TM CART5: a novel dual population CD5 CRISPR-Cas9 knocked out anti-CD5 chimeric antigen receptor T cell product for relapsed and refractory CD5+ nodal T-cell lymphomas. Blood 140(Supplement 1):1604–1605
Zhang J, Hu Y, Yang J, Li W, Tian Y, Wei G, Zhang L, Zhao K, Qi Y, Tan B, Zhang M (2020) Development and clinical evaluation of non-viral genome specific targeted CAR T cells in relapsed/refractory B-cell non-Hodgkin lymphoma. MedRxiv 23:2020–2009
Vuelta E, Ordoñez JL, Alonso-Pérez V, Méndez L, Hernández-Carabias P, Saldaña R, Sevilla J, Sebastián E, Muntión S, Sánchez-Guijo F, Hernández-Rivas JM (2021) CRISPR-Cas9 technology as a tool to target gene drivers in cancer: proof of concept and new opportunities to treat chronic myeloid leukemia. CRISPR J 4(4):519–535
Narimani M, Sharifi M, Jalili A (2019) Knockout of BIRC5 gene by CRISPR/Cas9 induces apoptosis and inhibits cell proliferation in leukemic cell lines, HL60 and KG1. Blood Lymphat Cancer: targets and therapy 27:53–61
Bexte T, Alzubi J, Reindl LM, Wendel P, Schubert R, Salzmann-Manrique E, von Metzler I, Cathomen T, Ullrich E (2022) CRISPR-Cas9 based gene editing of the immune checkpoint NKG2A enhances NK cell mediated cytotoxicity against multiple myeloma. Oncoimmunology 11(1):2081415
Naeimi Kararoudi M, Nagai Y, Elmas E, de Souza Fernandes Pereira M, Ali SA, Imus PH, Wethington D, Borrello IM, Lee DA, Ghiaur G (2020) CD38 deletion of human primary NK cells eliminates daratumumab-induced fratricide and boosts their effector activity. Blood 136(21):2416–2427
Chen X, Niu X, Liu Y, Zheng R, Yang L, Lu J, Yin S, Wei Y, Pan J, Sayed A, Ma X (2022) Long-term correction of hemophilia B through CRISPR/Cas9 induced homology-independent targeted integration. J Genet Genomics 49(12):1114–1126
Wang Q, Zhong X, Li Q, Su J, Liu Y, Mo L, Deng H, Yang Y (2020) CRISPR-Cas9-mediated in vivo gene integration at the albumin locus recovers hemostasis in neonatal and adult hemophilia B mice. Mol Ther Methods Clin Dev 11(18):520–531
Wang L, Yang Y, Breton CA, White J, Zhang J, Che Y, Saveliev A, McMenamin D, He Z, Latshaw C, Li M (2019) CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX–knockout mice. Blood J American Soc Hematol 133(26):2745–2752
Sung JJ, Park S, Choi SH, Kim J, Cho MS, Kim DW (2020) Generation of a gene edited hemophilia A patient-derived iPSC cell line, YCMi001-B-1, by targeted insertion of coagulation factor FVIII using CRISPR/Cas9. Stem Cell Res 1(48):101948
Park CY, Sung JJ, Cho SR, Kim J, Kim DW (2019) Universal correction of blood coagulation factor VIII in patient-derived induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rep 12(6):1242–1249
Author information
Authors and Affiliations
Contributions
The review topic was suggested by ZK and MM. The manuscript idea was reviewed and expanded by AA. Manuscript outlines were laid out by AA. Literature review was done by ZK, MM, AA, and FM. First draft was created by ZK and MM. Subsequent version was produced by AA. Revision and enhancement was done by FM.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abdulfatah M. Alayoubi and François E. Mercier are co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alayoubi, A.M., Khawaji, Z.Y., Mohammed, M.A. et al. CRISPR-Cas9 system: a novel and promising era of genotherapy for beta-hemoglobinopathies, hematological malignancy, and hemophilia. Ann Hematol 103, 1805–1817 (2024). https://doi.org/10.1007/s00277-023-05457-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-023-05457-2