Abstract
Island-mainland systems serve as natural laboratories for studying communication signals. We explored potential divergence in the song of the black-crowned antshrike between Gorgona Island, a land-bridge island off the Pacific coast of Colombia, and several mainland populations. We found that the perching song of this species, the so called loudsong, was distinctly different on Gorgona Island. This differentiation encompassed an increase in frequency bandwidth, a decrease in vocal performance, and a reduction in song duration. All are consistent with documented patterns of divergence observed in avian acoustic signals on oceanic islands. The observed distinctions in loudsong acoustics, including variations in frequency and temporal features, led to the inference that the population on Gorgona Island produces the least complex loudsong dialect across the entire species range. This pattern of how an innate vocalization manifests within a land-bridge island, which was connected to the mainland not long ago, lends support to the Character Release Hypothesis. To ensure an unbiased comparison, we considered genetic clusters and analyzed them by comparing ten acoustic signals between the land-bridge island and several mainland populations. Combined with the gene genealogy, this finding supports the validity of two geographic forms: Thamnophilus atrinucha atrinucha and Thamnophilus atrinucha gorgonae (island). However, they also reveal the existence of additional unrecognized dialects of loudsong that transcend current taxonomic classifications. We provide evidence that challenges the long-standing presumption suggesting minimal intraspecific geographic variation in the vocalization among non-vocal learning species.
Significance statement
Significant evolutionary implications exist for an intriguing pattern of variation of the loudsong behavior in the Black-crowned Antshrike over a land-bridge island. Our gene tree does provide evidence of the enormous haplotypic diversity accumulated in the trans-Andean region long after the final uplift of the northern Andes. The acoustic divergence of the loudsong suggests substantial geographic variation within a non-vocal learning suboscine. Genetic and loudsong divergence on a land-bridge island add to document that evolution of acoustic traits may be relaxed in islands as compared to those observed on the species-rich mainland. The island-mainland system is one of the few definitive biogeographic contexts for understanding evolution of acoustic behavior.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the field of evolutionary biology, island-mainland systems worldwide serve as one of the few definitive biogeographic contexts for understanding the mechanisms behind species variation. Species inhabiting islands often display phenotypic differences in terms of body size and shape, color, vagility, diet, fecundity and life history traits, when compared to their closely related counterparts on the mainland (Whittaker and Fernandez-Palacios 2007; Losos and Ricklefs 2009; Baeckens and van Damme 2020). One significant observation to highlight is that islands are characterized by lower species richness, making them species-poor communities compared to the mainland (Losos and Ricklefs 2009). This provides the context for any explanatory framework researching on how isolation can influence various traits. Based on this, it would be expected that the removal of a local species would lead to a reduced interspecific competition, forming the foundation for the Character Release Hypothesis (Grant 1972). Hence, in avian communities with a decline in richness, decreased interspecific competition, as proposed by Naugler and Ratcliffe (1994), open new ecological opportunities for the expansion and manifestation of traits, such as acoustic behavior on islands (Robert et al. 2019).
The study of avian acoustic behavior has become a valuable source of data for exploring the ecological and evolutionary patterns linked to vocal communication signals. Vocalizations are instrumental for individual recognition within species (Seddon 2005), a factor that can influence mate choice (Catchpole and Slater 1995). Consequently, bird song structure contains a substantial amount of inheritable phenotypic information (McCracken and Sheldon 1997). Among non-vocal learning species in passerines (suboscines), there is scant evidence of learning in the song development of specific species, particularly within Tyranni (Kroodsma 1984; Kroodsma et al. 2013) and Thamnophilidae (Touchton et al. 2014). Empidonax flycatchers' research has established the presence of a rudimentary neural-respiratory circuit that is far more developed than within vocal learning birds (Liu et al. 2013) and unveiled candidate genes showcasing differential expression in the suboscine brain of an Empidonax flycatcher related to bird song production (García et al. 2023). This suggests that the role of learning cannot be entirely dismissed in the acoustic behavior of non-vocal learning suboscines. Nevertheless, it remains evident that a substantial amount of data is encoded within what appear to be innate features of avian acoustics in non-vocal learning suboscines.
Non-vocal learning birds are typically expected to exhibit limited variation in their acoustic signals within species (Lindell 1998; Bard et al. 2002; Seddon and Tobias 2006; Tobias et al. 2010). Interestingly, geographic variation in vocalizations may function as localized communication signals for intraspecific discrimination. Moreover, the geographic structure of these vocalizations could likely be influenced by natural selection (Wilkins et al. 2013; Shipilina et al. 2017). Therefore, investigating the geographic variation in the songs of non-vocal learning birds has the potential to provide insight into understanding how populations' acoustic behavior diverges. Nevertheless, the absence of genetic data in studies of acoustic behavior may often hinder attempts to understand the spatial arrangement of songs in non-vocal learning birds (Sementili-Cardoso et al. 2018). Consequently, the spatial distribution of inherent vocal signals serves as a hypothesis regarding the geographic arrangement of genetic variation in suboscines.
The patterns of isolation within island biota create a context that is crucial to consider when interpreting the intricate dynamics of species' life cycles (Bellemain et al. 2008). Such patterns may indeed play a significant role in the rapid accumulation of functional diversity on islands (Theron et al. 2001). Both oceanic and land-bridge islands display remarkable patterns of endemism, suggesting the intriguing possibility that this endemism could also extend to communication signals. However, the accumulation of endemism in land-bridge islands may exhibit fundamental differences. These islands were once connected to the coastline, hosting taxa assemblages remarkably similar to those of the mainland. The process of accumulating biota on these islands is closely linked to their isolation due to rising sea levels (Han et al. 2022). This likely resulted in significant changes in the islands' relationship with the coastline and their biota (Wilson et al. 2015). Conversely, taxon assemblages on oceanic islands likely arose through long-distance dispersal, limited to highly vagile species, with only one or few colonization events shaping the circumstances for main evolutionary outcomes (Whittaker and Fernandez-Palacios 2007). Although the small area of oceanic landmasses makes these islands remarkable natural laboratories (Whittaker et al. 2017), it is important to note that near-shore islands are also significant biogeographical settings for the study of ecology and evolutionary biology (Wilson et al. 2015; Salerno et al. 2022).
Whether insularity affects behavior has received far less attention in the literature (Gavriilidi et al. 2022). In the context of the Character Release Hypothesis, several studies have documented distinctions in acoustic behavior when comparing avian species in oceanic islands and closely related counterparts on the mainland (Morinay et al. 2013; Robert et al. 2019, 2021, 2022), prompting our interest in this phenomenon. These studies suggest the release of acoustic space for sound-producing populations in oceanic islands, which results in that the remaining species on the islands will expand some of their acoustic traits in a less competitive environment compared to the mainland.
The Black-crowned Antshrike is a suboscine species in the suborder Tyranni that has shown to be a remarkable system for studying behavioral ecology (Tarwater 2012; Tarwater et al. 2013; de Zwaan and Roitberg 2015). Its range spans from Central America to the Northern Andes, encompassing elevations from the sea level to 1500 m (Brumfield and Edwards 2007). This species has a rather simple and stereotyped pattern of acoustic elements; a perching display of a recurrent loud patterned trill that starts slowly and culminates in a memorable terminal trill note, referred to as loudsong (Isler et al. 1997). In addition, T. atrinucha also emits rapid calls consisting of short repetitive sequences, often during low volume exchanges between paired individuals in close proximity, which are termed rattles (Isler et al. 1997). While these vocalizations likely serve various purposes, in the case of T. atrinucha, their specific functions as acoustic phenotypic traits in this species remain largely unknown. It is widely suggested that the loudsong may play a significant role in the species recognition system of thamnophilids (Isler et al. 1997, 1998, 2012).
In this study, we investigated the evolution in the frequency and temporal characteristics of the acoustic behavior and the genetic variation of the Black-crowned Antshrike in the island-mainland system of Gorgona-Colombia. Gorgona is a land-bridge island located 35 km off the Colombian Pacific coastline, which harbors a remarkable pattern of endemism (Fernández and Schaefer 2005; Giraldo et al. 2014). Two allopatric subspecies of the Black-crowned Antshrike, identified by subtle plumage coloration patterns, have been documented. T. atrinucha gorgonae, is presumed to be confined to Gorgona Island's rainforest (Thayer and Bangs 1905). T. atrinucha atrinucha, as suggested by Isler et al. (1997), is believed to inhabit tropical dry forests in Central America, extending from the Caribbean plain to Venezuela, including the inter-Andean valleys of Colombia. The specific form inhabiting the Pacific plain of Colombia and Ecuador (Chocó) remains unclear. Determining the distribution of each geographic species within the entire range is still pending, particularly concerning information on bird songs or genetic data.
Here we used song recordings and mitochondrial DNA from fieldwork conducted in previous years to investigate the geographic, genetic and acoustic patterns of this species. In addition, the potential influence of marginal learning on vocalizations suggests that we may anticipate some degree of relatable patterns of song and genetic divergence. Although our genetic dataset is small, it does offer a valuable independent set of data for further comparison with vocalizations. Mitogenomic data is a valuable tool for exploring the possible influence of vicariant events (Zink and Barrowclough 2008). It has also proven to be helpful in delineating boundaries within avian taxa (Gutiérrez-Zuluaga et al. 2021), including well-established Thamnophilus species (Thom and Aleixo 2015; Bolívar-Leguizamón et al. 2020). Given the challenges in objectively diagnosing certain species within the Thamnophilus genus based solely on plumage characteristics (Isler et al. 1997; Whitney et al. 2004; Bolívar-Leguizamón et al. 2020), the taxon under examination here may be polytypic. Thus, by integrating multiple types of data related to acoustic behavior, morphological traits, environmental factors and genetic data, we aim to examine a population in the 23-km2 insular landmass, which constitutes an ideal setting for studying the evolution of behaviors and communication signals.
Methods
Study area
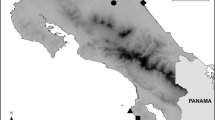
We collected vocalization recordings and tissue samples to obtain genotypic data for T. atrinucha in Colombia. Gorgona Island (2°9' N and 78°2' W) is 8.5-km long and 2.5-km wide and has a maximum elevation of 338 m and constitutes the primary study area to compare with mainland areas of Central America and Colombia (Fig. 1). The geographic range of T. atrinucha spans a diverse array of environments, encompassing elevations that vary in annual rainfall ranging from 1.5 to 6.7 m per year. It is evident that the species' habitat exhibits a high degree of complexity throughout the entire range.
Recordings of songs were obtained from locations in close proximity to the sites where specimens for genotyping were collected. Specific areas in Colombia (CO) are denoted by numbers: 1-Gorgona Island (GO), 2-Guapi (GU), 3-Nariño (NA), 4-Quindío (QU), 5-Antioquia (CA), 6-Cundinamarca (CU), 7-Choco (CHO), and for 8-Panama (PA), 9-Nicaragua (NI), and 10-Belize (BZ). However, no song recordings were available from BZ, and DNA sequences from NI were also absent. Sequence data for the ND2 gene were sourced from GenBank for BZ and PA, while data for CO were sequenced de novo
Collection and sampling
In 2016 and 2020, we used 30 mm mesh mist nets to collect individuals of T. atrinucha; A 10-µL blood sample was obtained through micro-puncture of the brachial vein, which we preserved in Queen's lysis buffer (Seutin et al. 1991). Additionally, during the preparation of skin specimens, we extracted approximately 3 mm3 of liver, heart, and muscle´s tissue. These tissue samples were then preserved either in denatured ethanol or lysis buffer. Five samples from Colombia were generously loaned to us by the avian collection of Universidad de Nariño and the Alexander von Humboldt Institute, respectively. Given the requirement to obtain all available specimens for our study, incorporating tissue samples blindly was impractical. However, to minimize observer bias, we implemented a blinded approach when recording and analyzing the acoustic behavior of focal animals in the field. The list of specimens used in this study, location and their GenBank accession numbers can be found in Fig. 2 and Supporting Online Information Table S1.
Consensus gen tree obtained from the phylogenetic inference based on partial sequences of the ND2 gene of Thamnophilus atrinucha. The nodal support is shown as the posterior probability from the Bayesian inference and the percent bootstrap value for 5,000 samples from the maximum likelihood analysis. The tip nodes list provides the GenBank accession number and specific areas with the country code: Colombia (CO), 1-Gorgona Island (GO), 2-Guapi (GU), 3-Nariño (NA), 4-Quindío (QU), 5-Antioquia (CA), 6-Cundinamarca (CU), 7-Choco (CHO), and for 8-Panama (PA), 10-Belize (BZ), and an unknown area (Unkn). The visualization of the posterior distribution uncertainty in a cloud diagram (green). Loudsong dialects next to the tree, a sample of vocal variation among subclades, the heat map towards warmer colors suggests greater intensity of energy emitted. Distribution of T. atrinucha and color code for an approximation of the spatial distribution of subclades. Sequences of the ND2 gene obtained from GenBank (BZ and PA) and sequence data obtained de novo (CO). T. atrinucha gorgonae, photo courtesy of NPA
DNA extraction and PCR
We conducted a characterization of mitochondrial DNA NADH dehydrogenase subunit 2 gene sequences as a preliminary approach to investigate the geographic genetic structure in T. atrinucha. To achieve this, we utilized samples collected from a substantial portion of its range in Colombia, and we also obtained additional sequences representing populations in Central America from the GenBank. We performed genomic DNA extractions using the Thermo Scientific® package and then further produced amplicons of the mtDNA ND2 gene. The PCR used previously described primers (Brumfield and Edwards 2007) and its product separated by electrophoresis on a 2% agarose gel with Safe DNA Dye (Hydragreen™). Amplicons of the expected size (ca. 1 Kb) were then sent to Macrogen Inc., for purification and sequencing. Sequencing of the light and heavy strands of mtDNA was conducted using BigDye (Applied BiosystemsTM). We aligned the sequences with Sequencher version 4.7 (Gene Codes Corporation, Ann Arbor, MI, USA), by using conspecific sequences from Panama and Belize obtained from GenBank (Benson et al. 2012).
Genealogical inference
We performed a genealogical analysis using a dataset comprising 30 ND2 partial sequences from Gorgona Island and the mainland. Most of these sequences were collected in Colombia (n = 23), while the remainder were sourced from GenBank, including sequences from Belize (n = 3) and Panama (n = 4). We performed a maximum likelihood (ML) inference analysis using the PhyML 3.0 platform (Guindon et al. 2010). The selection of the nucleotide substitution model was carried out using MEGA-X (Kumar et al. 2018). A total of 5,000 samples were collected from the inference to calculate the percentage support for the tree nodes. The gene tree inference included sequences from T. bridgesi (EF030293) and T. bernardi (EF030285) as external groups (Brumfield and Edwards 2007).
We also performed a Bayesian inference (BI) analysis using BEAST v1.10.4 (Suchard et al. 2018). The same substitution model was implemented in the Bayesian inference (BI) and ML analysis. To prepare the sequence file for BI analysis, we used BEAUTi v1.10.4 (Suchard et al. 2018). We configured a coalescent model with a constant population size, segregating the data into two partitions: one for codon positions 1 and 2 and another for position 3. During the analysis, data were sampled at intervals of 2000 steps from a total chain length of 200 million steps. We calibrated the genealogy based on the established monophyly reported in prior studies for T. atrinucha, T. bridgesi, and T. bernardi (Brumfield and Edwards 2007). We enforced a strict molecular clock and applied a rate of evolution of 0.029 substitutions per million years to the ND2 gene (Lerner et al. 2011).
After completion of the BI analysis, we inspected the resulting posterior estimates in Tracer (Rambaut et al. 2018), and excluded 25% of the initial sampled data as Burn-in. A visualization of the posterior distribution of sampled data after Burn-in was inspected using the Densitree 2 (Bouckaert and Heled 2014). We randomly selected 3 k trees from the stationary posterior distribution using R software (R Development Core Team 2019). These trees were subsequently used to construct a consensus majority tree using Paup v 4.0a (Swofford 2003). To assess the congruence between our Bayesian Inference (BI) and Maximum Likelihood (ML) analyses, we utilized node support as a reference criterion, requiring values of posterior probability ≥ 0.95 for BI and ≥ 60% bootstrap support for ML. Visualization of consensus tree was carried out using MrEnt v2.5 (Zuccon and Zuccon 2014).
Population genetic structure
The patristic (p) pairwise genetic distance was implemented to determine the nucleotide differences between ND2 gene sequences. We identified haplogroups among ND2 sequences using p-distances as implemented in USEARCH algorithm (Edgar 2010) with a nucleotide similarity criterion ≥ 99.99%, following the methodology outlined by Gil-Vargas and Sedano-Cruz (2019) and computed using MEGA-X (Kumar et al. 2018). We assessed the variation within T. atrinucha sequences by calculating nucleotide diversity (π), the number of parsimony-informative characters, and the Tajima D index (Tajima 1989) using Arlequin v.3.5.2.2 (Excoffier and Lischer 2010). We conducted the molecular analysis of variance (AMOVA) to estimate the nucleotide variation fixation index (Φst), using 16,000 permutations (Excoffier and Lischer 2010). Utilizing the matrix of Φst values as a representation of a neutral marker among different geographic areas, we explored whether the genetic clusters observed were influenced by geographical distance (Bohonak and Roderick 2001). The distance among sampling localities was calculated using the Geographic Distance Matrix Generator v1.2.3 (Ersts 2014).
Phenotypic characterization
Collection of vocalizations recording
We sourced audio recordings from both publicly available and unpublished archives, currently housed within the HFR's inventory. These recordings featured songs from sites in close to, or from the very areas where we collected specimens for genotyping (Fig. 1, Table S2). In Colombia, we recorded sounds in WAV format at a sampling rate of 44,000 Hz and 16 bits using different equipment. Most of the recordings were done with an XM-55 directional microphone and TASCAM DR-22WL solid-state recorder but in the Cauca River Canyon, we employed a Sennheiser Me67/K6 microphone with a Marantz PMD661 recorder (Supporting Online Information Table S2).
We also examined the sounds of T. atrinucha from Panama and Nicaragua in Central America (loudsong recordings from Belize were not available), obtained from public repositories such as the Macaulay Library at the Cornell Lab of Ornithology and Xeno-Canto. The data from the Macaulay Library were originally in 16-bit, 44,000 Hz WAV format; therefore, we had to transform the Xeno-Canto songs into WAV format with the same characteristics. The use of compressed files, such as those mp3 in the Xeno-Canto repository, is a topic of debate. However, both Heath et al. (2021) and Araya-Salas et al. (2019) coincide in suggesting that audio file compression does not affect acoustic measurements but in those designed to measure extreme frequencies. At any rate, the use of compressed audio files has provided reliable results and conclusions in acoustic analysis (Mikula et al. 2021). To mitigate potential issues related to data compression, as noted in Araya-Salas et al. (2019), only type A and B recordings (loud and clear) were selected from Xeno-Canto, based on their quality rating system. Then, we visually inspected the recordings spectrograms for subsequent analysis. If the target signal was distinctly distinguishable from the background noise, it was then chosen for further analysis. All recordings were normalized before any acoustic analysis. In addition, each recording represented only one individual. We focused on the acoustic data analysis of the loudsong (Isler et al. 1997), which manifests as a vocalization of multiple notes repeated at regular intervals with a clear end note that differs from the preceding notes.
Song analysis
We analyzed the structure of each loudsong using Avisoft-SASLab Pro Software, v. 5.2.09 (Avisoft Bioacoustics, Berlin, Germany). To quantify the acoustic spectrum of each sample, we employed Avisoft's automatic signal selection mode and set a detection threshold of -33 dB. This threshold was chosen to create standardized sonograms based on each song's peak amplitude, reducing the likelihood of spurious results attributable to measurement artifacts (Brumm et al. 2017). Subsequently, we derived spectrum-based measurements for nine acoustic parameters: Note duration (Dur), number of notes in the loudsong (Notes), silent intervals between notes (Break), strophe duration (ST-D), maximum frequency (max), minimum frequency (min), frequency peak (peak), bandwidth (Bandwd), and entropy (Entropy). The latter makes it possible to calculate the information content of the song, described in terms of purity or tonality of the sounds, where 0 indicates tonal sounds and 1 indicates sounds that accumulate greater entropy. We also estimated the rate at which notes were produced, a composite parameter that results from dividing the notes on ST-D. The note rate and ST-D are two common indicators of avian vocal performance (see a review on Robert et al. 2022). Because the songs of the same individual in a recording usually present low variability or are identical, we calculated the average values of notes per song/individual ST-D (sequence of loudsongs in a recording) for subsequent analyses.
To compare the pattern of acoustic structure between these groups of interest (e.g. regions or clades), we used the Wilcoxon test, which allowed us to examine the differential distribution pattern of the acoustic signal parameters. For the Wilcoxon test, we used scaled data per song per individual. Averaged values were calculated for each locality and arranged in pairs between groups of interest for further analysis. We also performed a discriminant analysis to reduce the dimensionality among the first nine parameters used to describe the structure of loudsong and thus, to maximize the acoustic discrimination between spatial groupings.
We calculated mean values per acoustic variable/locality or area of interest. The mean values were obtained from the acoustic spectrum of the loudsongs from all individuals sampled. Then, we calculated the Euclidean distance between pairs of areas of interest as a measurement of dissimilarity in a multidimensional space. The Euclidean distance is defined here as the root of the squared difference between the values of any pair of entities (Rivera-Gutierrez et al. 2010). Subsequently, we used this measurement of acoustic dissimilarity in a correlation analysis with the geographic and genetic distance matrices using the Cochran–Mantel–Haenszel test. All statistical analyses were performed in R software (R Development Core Team 2019).
Generalized dissimilarity analysis
We implemented a Generalized Dissimilarity Model (Ferrier et al. 2007), to estimate how environmental factors and geographic distance relate to the acoustic variation in loudsong of T. atrinucha. We utilized the GDM package for R (version 1.5) as described by Mokany et al. (2022) to employ the paired Acoustic Euclidean Distance as the response variable. This composite metric is founded on nine vocal structure parameters, excluding note rate. Four environmental variables were considered as predictors: BIO1 (Annual Mean Temperature), BIO7 (Temperature Annual Range, calculated as BIO5 Maximum annual temperature minus BIO6 Minimum annual temperature), BIO12 (Annual Precipitation) and a measure of vegetation cover (Leprieur et al. 2000): the Normalized Vegetation Index (NDVI) (Jiang and Huete 2010). Environmental data were extracted from Worldclim V. 2.1 (Fick and Hijmans 2017), using the Raster package for R software (Hijmans and van Etten 2023).
Body size analysis
Four measurements including exposed Culmen, Wing chord, Tarsus and Tail length, were used as proxies for body size. We conducted principal component analysis (PCA) for males and females to visually assess body size variation between island and mainland populations. In addition, we used the U-Mann Whitney comparison of body size measurements between areas of interest, such a non-parametric test was chosen due to the limited data available from the mainland. Specimens were obtained from the Universidad of Valle, Universidad de Nariño, the Institute of Natural Sciences from the Universidad Nacional of Colombia, and historical records from Gorgona Island (Thayer and Bangs 1905).
Phylogeographic and ecological clustering
We implemented the Bayesian phylogeographic and ecological clustering (BPEC, Manolopoulou et al. 2020) to examine whether differences in genetically distinct clusters are related to transitions of acoustic signals, body size and environmental variables. This model relies on the assumption that dispersal events result in the formation of new clusters, and these clusters are shaped by a multitude of factors, including the geographic distribution of haplotypes, phenotypic and ecological constraints on gene flow. We used the R-package BPEC to evaluate haplotype networks, jointly estimating genetic clusters along with georeferenced data points, nine acoustic variables, four male morphological traits derived from the prior body size analysis, and four environmental variables also used from the previous GDM analysis. Each dataset is represented in BPEC as the average observed at a specific location near the site where mtDNA haplotypes were collected. To run BPEC, missing sites in our sequence alignment were excluded. We also excluded three sequences from Belize to match only localities from where acoustic data of loudsongs were available. We performed pre-runs incrementing the number of dispersal events, and continued until maxMig + 1 yielded no change in the clustering output. Pre-runs included analysis after three, six and 10 million iterations, and relaxing haplotype network parsimony criteria ds from 0–3 (Manolopoulou et al. 2020).
Results
Genetic diversity
From mtDNA sequences for the ND2 gene (Supporting Online Information Table S2), we obtained an alignment of 1,008 base pairs (out of 1,042 bp -the total size of the ND2 gene in Thamnophilus), with an open reading frame without disruptive stop codons, and found no evidence of nuclear DNA copies. In this alignment, we found that the partial sequences of the ND2 gene had < 5% missing data and 96% were invariant loci, whereas 3% were informative loci (n = 28) (excluding outgroups). The mean number of pairwise differences is 6.2 nucleotides and averaged over loci π = 0.007. The p-distance among sequences was on average 1.1% (range 0.0 – 2.2%), which is a shorter distance than that found in other conspecifics (Lacerda et al. 2007). A total of 17 haplotypes were identified in the sequence alignment (Supporting Online Information Table S1).
The most common haplotype has a range distribution in Panama and Colombia, for the areas of Chocó, Nariño, and Gorgona Island (n = 12 sequences). We identified a single haplotype restricted to Panama and 14 haplotypes restricted to Colombia. The sequences from Belize (n = 3) formed a single haplotype group. The neutrality test for the entire set of haplotypes was not rejected (Tajima's D = − 1.31413; P = 0.086) implying an excess of low frequency polymorphism relative to the expected value.
Genealogical inference and dating
We used the alignment for the ND2 gene for the selection of a TN93 nucleotide substitution model (Tamura and Nei 1993). Maximum Likelihood (ML) inference showed strong support for the monophyly of T. atrinucha (log-likelihood score of -2005.64), as evidenced by a high bootstrap support. The ML analysis yielded results consistent with BI, as illustrated in Fig. 1. The divergence times for the ND2 gene in T. atrinucha can be traced back to the Pleistocene, with a broad confidence interval ranging from 0.03 to 4.88 million years ago. When considering the diversification of subclades with robust posterior probabilities and ML bootstrap support, it becomes evident that there was a significant overlap in node age at the 95% high posterior density interval (data not shown). Consequently, we lacked the statistical power to discern distinct divergence times among subclades.
Both the bootstrap from ML and the posterior probability (PP) estimation from BI supported a genealogy, which also highlights a geographically structured pattern characterized by three significant subclades (Fig. 2). The first subclade comprises haplotypes distributed within the westernmost area of the species' range, which includes areas in Belize, Panama, and the Pacific coast of Colombia, and the population found on Gorgona Island (Fig. 2). The second clade is composed of haplotypes distributed throughout the easternmost part of the inter-Andean region of Colombia (Eastern mountain range), hereafter Eastern region. The third clade comprises haplotypes distributed within Colombia's inter-Andean region, spanning from the lower Rio Cauca in Antioquia to the department of Quindío (Central mountain range of Colombia), hereafter Central region. Further, the spatial distribution of nucleotide variation was high (AMOVA Φst 1, 27 = 0.76601, Exact Test of individual distribution P = 0.00001), when contrasting the Pacific region (including the Gorgona Island´s population), with the broader Andean region of Colombia encapsulated by the Central and Eastern mountain ranges subclades.
Loudsong acoustic signals
The database comprised a total of 266 loudsongs gathered from 43 individuals of T. atrinucha. These recordings were made in the same geographical areas or nearby sites where genetic samples were collected for analysis. The loudsong lasted ≤ 2.5 s (Fig. 2). The Pacific region subclade displayed the longest and shortest songs in the entire dataset —Guapi and Gorgona Island, respectively— separated by just 60 km (Fig. 2). Despite the overall similar appearance of loudsongs across the species' entire range, distant populations likely exhibit differences in multiple acoustic parameters. However, it is difficult once in the field to objectively distinguish such differences in acoustics of the loudsong.
When comparing loudsongs within regions, particularly emphasizing the populations from Gorgona and Panama, distinctions emerged in most acoustic parameters (Fig. 3, Supporting Online Information Table S3). The acoustic signals of T. atrinucha from Gorgona Island showed differences in no fewer than seven parameters compared to other regions (Fig. 3, Supporting Online Information Table S4). In terms of performance, while the mean note duration fell within intermediate values when compared to other regions, birds from Gorgona Island showed the lowest number of notes and song rate, along with the longest breaks between notes. Additionally, birds from Gorgona Island sang at both the highest peak and maximum frequency, concurrently displaying one of the lower frequencies (Fig. 3, Supporting Online Information Table S4). This variance in frequency contributed to the broadest bandwidth in their acoustic signals in Gorgona Island (Fig. 3).
Regional comparison of Loudsong in Thamnophilus atrinucha among major subclades identified in the ND2 gene genealogy, specifically the eastern mountain range (Andes), the central range (Cauca), and the Pacific region. Moreover, the comparison offers detailed analyses of populations from Gorgona Island (T. atrinucha gorgonae) and Panama (T. atrinucha atrinucha), both situated within the Pacific region subclade (see Fig. 1). This comparison implement mean values and standard errors for each acoustic signal, encompassing the duration of the note (Dur), strophe duration (ST- D), number of notes in the strophe (Notes), frequency peak (peak), minimum frequency (min), maximum frequency (max), bandwidth (Bandwd), Entropy, and silence time between notes (Break)
Seven distinct parameters within the acoustic signal of T. atrinucha's loudsong provided a clear differentiation between the Central inter-Andean and Eastern mountain range regions. On average, loudsongs in the Eastern mountain range had lower acoustic frequencies (min, max, and peak), fewer notes, a generally reduced note rate, and a narrower bandwidth when compared to the songs from the Central mountain range region (Fig. 3, Supporting Online Information Table S4).
When comparing the loudsongs from the Pacific and Central inter-Andean regions, various key differences emerged. In the Central region, the maximum frequency of the song was notably lower, shorter in duration, notes were briefer, and, overall, the note rate was higher. Conversely, in the Pacific region, loudsongs tended to be longer and consist of more notes; however, the individual notes within the song were shorter in duration and were emitted at higher frequencies (peak and max.) (Fig. 3, Supporting Online Information Table S4). Despite these differences in the number of notes and strophe duration, both the Eastern mountain range and Pacific regions shared a similar note rate (Fig. 3, Supporting Online Information Table S4).
We used the spatial distribution of three major subclades in the ND2 gene tree to conduct a discriminant analysis of the loudsong. This analysis showed that the acoustic data was sufficient to describe the structure of loudsong in two discriminant functions of phylogeographic regions (df = 2; Wilks = 0.40658; approx. F22506 = 13.071; P < 0.001). These functions lower the dimensionality of nine acoustic signals (Supporting Online Information Fig. S1), and maximize the total variation observed between regions (subclades) established a priori (LD1 = 80.4%, LD2 = 19.6%). This resulted in the correct assignment of each song to its region of origin for 95% of the loudsongs in T. atrinucha. The discriminant pattern of the acoustic signals for the Central region (subclade) is located at a midpoint between the Eastern mountain range and the Pacific regions, which in turn do not overlap (Supporting Online Information Fig. S1).
Loudsong acoustics, genetic, geographic and environmental distances
Despite the approximate 600-km separation between some localities in the Pacific region, populations of Gorgona Island and Panama differed in all nine acoustic variables (Fig. 3, Supporting Online Information Table S3) and showed the closest genetic p-distance (0.09% ± 0.06). Conversely, with a greater genetic p-distance (0.21% ± 0.06) between Gorgona Island and mainland areas of Chocó, Guapi, and Nariño in the Pacific coast of Colombia, acoustic differences were evident, including seven variables (Fig. 3, Supporting Online Information Table S3). Notably, geographic distances among populations in the Pacific region of mainland Colombia and the land-bridge island span a shorter range of 60 to 300 km. It's noteworthy that six distinct acoustic signals effectively differentiated the populations in Panama from those in the mainland regions of the Pacific coast in Colombia. It's also important to emphasize that when comparing these mainland populations to Panama, a higher genetic p-distance of 0.21% ± 0.05 is observed, contrasting with the higher genetic similarity previously noted between Gorgona Island and Panama. Geographic and environmental distances explained roughly 27.3% of the variance on the acoustic Euclidean distance, according to the GDM analysis. The I-Spline coefficients showed that Temperature Annual Range (BIO7) and Geographic distance played the largest role explaining acoustic distance (BIO7: 0.524; Geographic distance: 0.427). In simple terms, the I-Spline coefficient represents the minimum "force" required to determine the best-fitted polynomial curve for each factor as a function of acoustic distance. While the directionality of acoustic response is a rather complex effect, all coefficients including those with a lower I-Spline value (NDVI: 0.165; BIO1: 0.159 and BIO12: 0.135) suggest to us that bioclimatic variables and vegetation structure, all contributed to explain the spatial variation in acoustic characteristics of the loudsong in T. atrinucha.
Body size between Gorgona Island and Mainland
We obtained body measurement data for 56 males and 48 females of T. atrinucha, of which 16 and 11 corresponded to males and females from Gorgona Island. The principal component analysis can account for as much as 37.3% of the variation in the four morphological measurements for males and 41.1% for females (Supporting Online Information Fig. S2). Males and females from the island tended to have larger bodied-size when compared to mainland populations from the Pacific and inter-Andean regions. Females in Gorgona Island had longer wing length, tarsi, and tails when compared to mainland (p < 0.05 for each variable among the available specimens). Male specimens from the island consistently showed larger culmen, wings, tarsi, and tails when compared to multiple mainland locations (p < 0.02 for each variable among the available specimens). When comparing male specimens from Gorgona Island, which is only 60 km away from Guapi, an evident difference in exposed culmen length becomes apparent. In contrast, for females, the variation in culmen length was more subtle, indicating a marginal level of difference (p < 0.053).
Phylogeographic, phenotypic and ecological clustering
The parsimony network obtained using BPEC supports three geographically distinct clusters of T. atrinucha (PP = 1.0) (Fig. 4, Supporting Online Information Fig. S3). The spatial distinctiveness among these genetic clusters varies significantly and was influenced by nine acoustic variables, four morphological measurements for males, three bioclimatic variables, and data pertaining to vegetation structure. The first cluster clearly comprised haplotypes sampled from Gorgona island, Guapi and Nariño (SW-Pacific Colombian coast). The second cluster was composed of haplotypes with distribution in Panama and northern Chocó. The third was an Andean cluster that comprises haplotypes from Central and Eastern mountain ranges of Colombia (Fig. 4, Supporting Online Information Fig. S3). Nonetheless, BPEC strongly suggested that it is difficult to separate the latter from the Panama and the northern Chocó cluster (PP = 0.64). The transitions between clusters were driven by differences in means of the acoustic, body size and environmental variables rather than their variances. The modulation of acoustic variation reveals a trend that is not strictly unidirectional, as shown by variation in spatial transitions among clusters in the nine acoustic signals of the loudsong. According to BPEC the parsimony network lacked any loops to be resolved, which may lead to equivocal connections between haplotypes (Manolopoulou et al. 2020). However, the clusters do not correspond strictly with the spatial distribution of the primary subclades identified in the phylogenetic inference (Fig. 2, 4). This outcome discrepancy was unsurprising, as it aligns with the inherently distinct approach of integrating multiple data types within BPEC.
Distribution of covariates for each phylogeographic cluster pertaining to Thamnophilus atrinucha dataset using BPEC. The colors correspond to the same clusters depicted in the contour map (top left panel). Panels show spatial transitions involving acoustic signals, body size, and environmental variables among clusters. The shaded area correspond to the 5% and 95% credibility intervals for each cluster, while the solid lines denote the median for each point. The presented subset comprises two acoustic signals, Note and Bandwidth (Bandwd), in addition to measurements of exposed Culmen and Wing chord for males, along with Temperature Annual Range. A comprehensive set of 17 variables were implemented in the full BPEC analysis (Supporting Online Material Fig. S3). This set included nine acoustic parameters, four morphological traits, four remote sensing variables associated with environmental factors, and an index of vegetation structure
Discussion
Our study on the variation in loudsong acoustic signals covered the entire range of a non-vocal learning Thamnophilid species. In particular, we found evidence of evolutionary changes in loudsong between island and mainland populations within T. atrinucha. In this species the mtDNA ND2 gene form a monophyletic group and exhibit non-paraphyletic relationships with T. bridgesi and T. bernardi (external groups), contributing to a more comprehensive understanding of the genus Thamnophilus (Brumfield and Edwards 2007). While we acknowledge the limitations associated with relying solely on a single maternally inherited genetic marker for drawing conclusions about the genealogical process (see a study case in Palacios et al. 2023), our genetic data, at the very least, provides a supportive genealogical framework for analyzing the acoustic variation in loudsongs between Gorgona Island and mainland populations.
We documented clear differences in acoustic variables of T. atrinucha using as a line of reference three well-supported mtDNA lineages in the discrimination analysis (Supporting Online Information Fig. S1). On the contrary, Bayesian phylogeographic and ecological clustering does not involve reducing the dimensionality of multiple acoustic signals into discriminant functions. It also does not depend on any preconceived assumptions about spatial groupings, as is the case in a discriminant function analysis. The BPEC analysis reveals spatially structured clusters based on genetic, geographic, acoustic, morphological and environmental data. We highlight the observed shifts in the relative significance of individual variables across space (Fig. 4, Supporting Online Information Fig. S3). This underscores the complex spatial relationship between song structure and the varying contribution of each variable, a connection previously identified through the Generalized Dissimilarity Model.
Our integrative approach of multiple types of data was instrumental for the phylogeographic and ecological clustering of T. atrinucha. These clusters imply an unambiguous pattern of divergence if one considers that differences in two or more characteristics of the acoustic signals as sufficient criteria to distinguish between species of the genus Thamnophilus (Isler et al. 1997, 1998, 2012) and in other avian groups (Cadena and Cuervo 2010; Lara et al. 2012). Here, the distributional range of T. a. gorgonae is being extended beyond Gorgona Island to nearby areas along the SW-Pacific Colombian coast. According to our BPEC analysis, there is support for the unambiguous differentiation of T. a. atrinucha in Panama from T. a. gorgonae. From our results, the ND2 gene tree, acoustic, body size and environmental data strongly imply that a polytypic classification (Clements et al. 2016; IOC version 7.2, 2017) divided only into T. a atrinucha (Salvin and Godman 1892) and T. a. gorgonae (Thayer and Bangs 1905) appears insufficient. These two well differentiated geographic populations are part of the same subclade within the Pacific region and Central America, but additional subclades in the gene tree minimally imply that there are likely unrecognized populations elsewhere. It is worth recognizing that the number of samples for some areas was smaller than we would have intended. Nevertheless, we emphasize that our primary goal was to investigate the hypothesis of population divergence in Gorgona Island; we believe that the size of our sample adequately served its intended purpose.
Loudsong structure within the Pacific region, where T. atrinucha in Gorgona Island belongs to, does not seem restricted in terms of their genetic distance for the loss or gain of signals in their acoustic repertoire (Fig. 4). Our main finding is that two vocal attributes of loudsong in T. atrinucha inhabiting a land-bridge island are consistent with predicted patterns of avian song divergence between oceanic islands and mainland (Robert et al. 2021, 2022). The Gorgona Island population with larger bodied-size individuals produces a loudsong with the largest increase in frequency bandwidth of acoustic signals and the lowest note rate. Here, birds´ loudsong manifestation appears relaxed showing the lowest performance (song rate), which is achieved through the longest breaks between notes.
Because the number of notes is a phylogenetically informative trait in species with unlearned vocalizations (McCracken and Sheldon 1997; Carneiro et al. 2019); we argue that the lowest number of notes (but longer notes) of T. atrinucha in Gorgona Island is a reasonable proxy to describe the species´ simplest stereotyped loudsong. Although the description of complexity for vocal phenotypes is a topic still under discussion and nowhere near a consensus (Pearse et al. 2018; Najar and Benedict 2019); the diversity of notes has been proposed to be an informative variable when the number and variety of the notes are considered (Wolf 1977). The lowest note rate and the short duration of the loudsong of T. atrinucha in Gorgona Island add to the notion that the acoustic structure in the island´s endemic is the least complex Loudsong, in support of the argument of a relaxed vocal performance in the island.
Abiotic and biotic context to Loudsong
Regarding local restrictions, it is reasonable to speculate that environmental heterogeneity, such as variations in rainfall, temperature, or habitat structure, could influence not only the structure of avian communities (Boncoraglio and Saino 2007; Brown et al. 2017), but also the aspects related to the effectiveness of acoustic communication. Our GDM and BPEC analyses both indicate that bioclimatic variables, vegetation structure, and geographic distance may significantly influence the spatial acoustic variation of T. atrinucha. The shaping of acoustic variation among clusters exhibits a non-unidirectional trend, evident in spatial transitions across nine acoustic signals of the loudsong. Specifically, in populations along the southwest Pacific coast of Colombia (including Gorgona Island), the increased frequency bandwidth of acoustic signals, when compared to other clusters, may be linked to transitions toward lower annual temperature ranges (BIO7) and reduced measurements of vegetation cover (NDVI) (Supporting Online Material Fig. S3). How these and other biotic factors relate to loudsong divergence awaits further research. Subsequent studies may examine if the increased frequency bandwidth of acoustic signals is mediated by environmental restrictions or local noise near the shore on the 23-km2 island. An increase in the frequency of sound emission in birds has been suggested as an adaptive strategy for avoiding acoustic interference caused by environmental constraints, including ocean noise (Sebastianelli et al. 2021) or perhaps the omnipresent sound of insects, which are an important generator of noise in tropical rainforests (Luther and Gentry 2013).
However, even when the acoustic signal of T. atrinucha is not distorted by a rainier environment, the acoustic interference from other species in the community may be different and may promote acoustic acclimatization (Luther 2009) or song evolution to reduce interference. A forest depleted of bird sounds characterizes Gorgona Island compared with the loud choruses of birds in the forest by the coast, home to 20 times richer avian fauna than that found on the island (RES-C pers. obs.). In a context such as this, birds on the island would have the opportunity to emit vocalizations using a less demanding portion of the acoustic spectrum (Torres et al. 2020). To us, the loudsong phenomena in Gorgona Island would be consistent with acoustic relaxation of song, in support of the character release hypothesis for avian vocalizations in islands (Robert et al. 2021).
Changes in the area of Gorgona Island and its relationship with the coastal line as a result of recurrent changes in the sea level cannot be discounted considering that only 35 km of maritime gap separates the mainland. The areal reduction would possibly promote the restricted scattering or cycles of local isolation as geographic permeability for dispersal had been suggested to influence the range limits of many birds of the northern Andes (Graham et al. 2010; Valderrama et al. 2014). There are also important characteristics of T. atrinucha that presumably relate to the observed pattern presented here, including high dwelling fidelity and low post-fledging dispersal (Tarwater 2012; Tarwater et al. 2013) and the possible restriction to dispersal by specialization as documented in other species of the genus Thamnophilus (Choueri et al. 2017). All these factors would lead to a historical pattern of differential accumulation of acoustic signals and of neutral genetic variation as documented here in a non-vocal learning suboscine.
It is tempting to hypothesize that local divergence of loudsong in Gorgona Island has been promoted primarily in isolation. The local effect of a reduction in the acoustic interference of the bird community cannot be ruled out (Capelli et al. 2020; Torres et al. 2020). We suggest field experiments to examine whether the acoustic divergence of T. atrinucha may be coupled with a rapid discrimination of communication signals from the mainland and the selection phenomenon for better signal transmission (Barker 2008). Acoustic divergence may also avoid acoustic interference within a lineage that share the same habitat type (Slabbekoorn and Smith 2002; Cicero et al. 2020), perhaps by a subtle adjustment of the signals during the crystallization of the song (Touchton et al. 2014). Even slight variations in innate song can contribute to the specific recognition (Macedo et al. 2019). Innate vocalizations are expected to have slowly accumulated small mutations in response to local adaptation (Laiolo and Rolando 2002; Nicholls and Goldizen 2006; Tobias et al. 2010). Our results collectively suggest that invoking neutral evolution of the loudsong in the light of our genetic, acoustic, body size and environmental data seems insufficient to explain the geographic variation of loudsong among closely related populations of T. atrinucha.
Data availability
The authors declare that all data supporting the findings of this study are available within this article [and its supplementary information files].
References
Araya-Salas M, Smith-Vidaurre G, Webster M (2019) Assessing the effect of sound file compression and background noise on measures of acoustic signal structure. Bioacoustics 28:57–73. https://doi.org/10.1080/09524622.2017.1396498
Baeckens S, Van Damme R (2020) The island syndrome. Curr Biol 30:R338–R339. https://doi.org/10.1016/j.cub.2020.03.029
Bard SC, Hau M, Wikelski M, Wingfield JC (2002) Vocal distinctiveness and response to conspecific playback in the Spotted Antbird, a neotropical suboscine. Condor 104:387–394
Barker NK (2008) Bird song structure and transmission in the neotropics: trends, methods and future directions. Ornitol Neotrop 19:175–199
Bellemain E, Bermingham E, Ricklefs RE (2008) The dynamic evolutionary history of the bananaquit (Coereba flaveola) in the Caribbean revealed by a multigene analysis. BMC Evol Biol 8:240. https://doi.org/10.1186/1471-2148-8-240
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2012) GenBank. Nucleic Acids Res 41:D36–D42. https://doi.org/10.1093/nar/gks1195
Bohonak AJ, Roderick GK (2001) Dispersal of invertebrates among temporary ponds: are genetic estimates accurate? Isr J Zool 47:367–386
Bolívar-Leguizamón SD, Silveira LF, Derryberry EP, Brumfield RT, Bravo GA (2020) Corrigendum to “Phylogeography of the Variable Antshrike (Thamnophilus caerulescens), a South American passerine distributed along multiple environmental gradients” [Mol Phylogenet Evol 148 (2020):106810. 10.1016/j.ympev.2020.106810]. Mol Phylogenet Evol 150:106890
Boncoraglio G, Saino N (2007) Habitat structure and the evolution of bird song: a meta-analysis of the evidence for the acoustic adaptation hypothesis. Funct Ecol 21:134–142. https://doi.org/10.1111/j.1365-2435.2006.01207.x
Bouckaert R, Heled J (2014) DensiTree 2: seeing trees through the forest. bioRxiv. https://doi.org/10.1101/012401
Brown PT, Ming Y, Li W, Hill SA (2017) Change in the magnitude and mechanisms of global temperature variability with warming. Nat Climate Change 7:743–748. https://doi.org/10.1038/nclimate3381
Brumfield RT, Edwards SV (2007) Evolution into and out of the Andes: a Bayesian analysis of historical diversification in Thamnophilus antshrikes. Evolution 61:346–367. https://doi.org/10.1111/j.1558-5646.2007.00039.x
Brumm H, Zollinger SA, Niemelä PT, Sprau P (2017) Measurement artifacts lead to false positives in the study of birdsong in noise. Methods Ecol Evol 8:1617–1625. https://doi.org/10.1111/2041-210X.12766
Cadena CD, Cuervo AM (2010) Molecules, ecology, morphology, and songs in concert: how many species is Arremon torquatus (Aves: Emberizidae)? Biol J Linn Soc 99(1):152–176. https://doi.org/10.1111/j.1095-8312.2009.01333.x
Capelli D, Batalha-Filho H, Japyassú HF (2020) Song variation in the Caatinga suboscine Silvery-cheeked Antshrike (Sakesphorus cristatus) suggests latitude and São Francisco River as drivers of geographic variation. J Ornithol 161:873–884. https://doi.org/10.1007/s10336-020-01779-4
Carneiro L, Bravo GA, Aleixo A (2019) Phenotypic similarity leads to taxonomic inconsistency: A revision of the lowland’s antpittas. Zool Scr 48:46–56. https://doi.org/10.1111/zsc.12324
Catchpole CK, Slater PJB (1995) Bird Song: Biological themes and Variations. Cambridge University Press, Cambridge
Choueri ÉL, Gubili C, Borges SH, Thom G, Sawakuchi AO, Soares EAA, Ribas CC (2017) Phylogeography and population dynamics of Antbirds (Thamnophilidae) from Amazonian fluvial islands. J Biogeogr 44:2284–2294. https://doi.org/10.1111/jbi.13042
Cicero C, Mason NA, Benedict L, Rising JD (2020) Behavioral, morphological, and ecological trait evolution in two clades of New World Sparrows (Aimophila and Peucaea, Passerellidae). PeerJ 8:e9249. https://doi.org/10.7717/peerj.9249
Clements JF, Schulenberg TS, Iliff MJ, Roberson D, Fredericks TA, Sullivan BL, Wood CL (2016) The eBird/Clements checklist of birds of the world. The Cornell Lab Ornithology, Ithaca
de Zwaan DR, Roitberg BR (2015) Variation in foraging strategy with forest age for the Black-Crowned Antshrike (Thamnophilus atrinucha). Ornitol Neotr 26:266–282
Edgar R (2010) Drive5. http://drive5.com/usearch/. Accessed 21 Jul 2023
Ersts PJ (2014) Geographic distance matrix generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation, New York. http://biodiversityinformatics.amnh.org/open_source/gdmg. Accessed 21 Mar 2022
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Fernández L, Schaefer SA (2005) New Trichomycterus (Siluriformes: Trichomycteridae) from an Offshore Island of Colombia. Copeia 2005:68–76. https://doi.org/10.1643/CI-04-177R1
Ferrier S, Manion G, Elith J, Richardson K (2007) Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Diver Distrib 13:252–264. https://doi.org/10.1111/j.1472-4642.2007.00341.x
Fick SE, Hijmans RJ (2017) Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas 2017. http://worldclim.org/version2. Accessed 12 Feb 2024
García NC, Campagna L, Rush AC, Bowie RCK, Lovette IJ (2023) Comparative genomics of two Empidonax flycatchers reveal candidate genes for bird song production. Evolution 77:1818–1828. https://doi.org/10.1093/evolut/qpad096
Gavriilidi I, De Meesters G, Van Damme R, Baeckens S (2022) How to behave when marooned: the behavioural component of the island syndrome remains underexplored. Biol Lett 18:20220030. https://doi.org/10.1098/rsbl.2022.0030
Gil-Vargas DL, Sedano-Cruz RE (2019) Genetic variation of avian malaria in the tropical Andes: a relationship with the spatial distribution of hosts. Malaria J 18:129. https://doi.org/10.1186/s12936-019-2699-9
Giraldo A, Diazgranados MC, Gutierrez-Landazuri CF (2014) Isla Gorgona, enclave estratégico para los esfuerzos de conservación en el Pacífico Oriental Tropical. Rev Biol Trop 62:1–12
Graham CH, Silva N, Velásquez-Tibatá J (2010) Evaluating the potential causes of range limits of birds of the Colombian Andes. J Biogeogr 37:1863–1875. https://doi.org/10.1111/j.1365-2699.2010.02356.x
Grant PR (1972) Convergent and divergent character displacement. Biol J Linn Soc 4:39–68. https://doi.org/10.1111/j.1095-8312.1972.tb00690.x
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Gutiérrez-Zuluaga AM, González-Quevedo C, Oswald JA, Terrill RS, Pérez-Emán JL, Parra JL (2021) Genetic data and niche differences suggest that disjunct populations of Diglossa brunneiventris are not sister lineages. Ornithology 138:ukab015. https://doi.org/10.1093/ornithology/ukab015
Han P, Zhao Y, Kang Y, Ding P, Si X (2022) Island biogeography of soundscapes: Island area shapes spatial patterns of avian acoustic diversity. J Biogeogr. https://doi.org/10.1111/jbi.14551. (published online)
Heath BE, Sethi SS, Orme CDL, Ewers RM, Picinali L (2021) How index selection, compression, and recording schedule impact the description of ecological soundscapes. Ecol Evol 11:13206–13217. https://doi.org/10.1002/ece3.8042
Hijmans RJ, van Etten J (2023) Raster: geographic data analysis and modeling (R package). https://CRAN.R-project.org/package=raster. Accessed 12 Feb 2024
Isler ML, Isler PR, Whitney BM (1997) Biogeography and systematics of the Thamnophilus punctatus (Thamnophilidae) complex. Ornithol Monogr 48:355–381
Isler ML, Isler PR, Whitney BM (1998) Use of vocalizations to establish species limits in antbirds (Passeriformes: Thamnophilidae). Auk 115:577–590
Isler ML, Cuervo AM, Bravo GA, Brumfield RT (2012) An integrative approach to species-level systematics reveals the depth of diversification in an Andean Thamnophilid, the Long-tailed Antbird. Condor 144:571–583. https://doi.org/10.1525/cond.2012.120012
Jiang Z, Huete AR (2010) Linearization of NDVI based on its relationship with vegetation fraction. Photogramm Eng Rem S 76:965–975. https://doi.org/10.14358/PERS.76.8.965
Kroodsma DE (1984) Songs of the Alder Flycatcher (Empidonax alnorum) and Willow Flycatcher (Empidonax traillii) are innate. Auk 10:13–24
Kroodsma DE, Hamilton D, Sánchez JE, Byers BE, Fandiño-Mariño H, Stemple DW, Trainer JM, Powell JVN (2013) Behavioral evidence for song learning in the suboscine bellbirds (Procnias spp.; Cotingidae). Wilson J Ornithol 125:1–14
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lacerda D, Marini M, Santos F (2007) Mitochondrial DNA corroborates the species distinctiveness of the Planalto (Thamnophilus pelzelni Hellmayr, 1924) and the Sooretama (T. ambiguus Swainson, 1825) Slaty-antshrikes (Passeriformes: Thamnophilidae). Braz J Biol 67:873–882. https://doi.org/10.1590/s1519-69842007000500010
Laiolo P, Rolando A (2002) The begging call of the Red-billed Chough Pyrrhocorax pyrrhocorax: an analysis of its spectrotemporal variability. Dissertation, University of Torino. http://hdl.handle.net/2318/8935. Accessed Mar 2024
Lara CE, Cuervo AM, Valderrama SV, Calderón-F D, Cadena CD (2012) A new species of wren (Troglodytidae: Thryophilus) from the dry Cauca River Canyon, Northwestern Colombia. Auk 129(3):537–550. https://doi.org/10.1525/auk.2012.12028
Leprieur C, Kerr YH, Mastorchio S, Meunier JC (2000) Monitoring vegetation cover across semi-arid regions: comparison of remote observations from various scales. Int J Remote Sens 21:281–300. https://doi.org/10.1080/014311600210830
Lerner HR, Meyer M, James HF, Hofreiter M, Fleischer RC (2011) Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr Biol 21:1838–1844
Lindell C (1998) Limited geographic variation in the vocalizations of a neotropical furnarid, Synallaxis albescens. Wilson Bull 110:368–374
Liu WC, Wada K, Jarvis ED, Nottebohm F (2013) Rudimentary substrates for vocal learning in a suboscine. Nat Commun 4:2082. https://doi.org/10.1038/ncomms3082
Losos JB, Ricklefs RE (2009) Adaptation and diversification on islands. Nature 457:830–836
Luther D (2009) The influence of the acoustic community on songs of birds in a neotropical rain forest. Behav Ecol 20:864–871. https://doi.org/10.1093/beheco/arp074
Luther D, Gentry K (2013) Sources of background noise and their influence on vertebrate acoustic communication. Behaviour 150:1045–1068
Macedo G, Silva M, Do AFR, Maldonado-Coelho M (2019) Symmetrical discrimination despite weak song differentiation in two suboscine bird sister species. Behav Ecol 30:1205–1215. https://doi.org/10.1093/beheco/arz066
Manolopoulou I, Hille A, Emerson B (2020) BPEC: an R package for Bayesian phylogeographic and ecological clustering. J Stat Softw 92:1–32. https://doi.org/10.18637/jss.v092.i05
McCracken KG, Sheldon FH (1997) Avian vocalizations and phylogenetic signal. P Natl Acad Sci USA 94:3833–3836. https://doi.org/10.1073/pnas.94.8.3833
Mikula P, Valcu M, Brumm H, Bulla M, Forstmeier W, Petrusková T, Kempenaers B, Albrecht T (2021) A global analysis of song frequency in passerines provides no support for the acoustic adaptation hypothesis but suggests a role for sexual selection. Ecol Lett 24:477–486. https://doi.org/10.1111/ele.13662
Mokany K, Ware C, Woolley SNC, Ferrier S, Fitzpatrick MC (2022) A working guide to harnessing generalized dissimilarity modelling for biodiversity analysis and conservation assessment. Global Ecol and Biogeogr 31:802–821. https://doi.org/10.1111/geb.13459
Morinay J, Cardoso GC, Doutrelant C, Covas R (2013) The evolution of birdsong on Islands. Ecol Evol 3:5127–5140. https://doi.org/10.1002/ece3.864
Najar N, Benedict L (2019) The relationship between latitude, migration and the evolution of bird song complexity. Ibis 16:1–12. https://doi.org/10.1111/ibi.12648
Naugler CT, Ratcliffe L (1994) Character release in bird song: a test of the acoustic competition hypothesis using American tree sparrows Spizella arborea. J Avian Biol 25:142–148. https://doi.org/10.2307/3677033
Nicholls JA, Goldizen AW (2006) Habitat type and density influence vocal signal design in satin bowerbirds. J Anim Ecol 75:549–558. https://doi.org/10.1111/j.1365-2656.2006.01075.x
Palacios C, Campagna L, Parra JL, Cadena CD (2023) Mito-nuclear discordance in the phenotypically variable Andean hummingbirds Coeligena bonapartei and Coeligena helianthea (Trochilidae). Bio J Linnean Soc 139:145–157. https://doi.org/10.1093/biolinnean/blad013
Pearse WD, Morales-Castilla I, James LS, Farrell M, Boivin F, Davies TJ (2018) Complexity is complicated and so too is comparing complexity metrics‐A response to Mikula et al. (2018). Evolution 72:2836–2838. https://doi.org/10.1111/evo.13636
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed 5 Mar 2024
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Rivera-Gutierrez HF, Matthysen E, Adriaensen F, Slabbekoorn H (2010) Repertoire sharing and song similarity between great tit males decline with distance between forest fragments. Ethology 116:951–960. https://doi.org/10.1111/j.1439-0310.2010.01813.x
Robert A, Lengagne T, Melo M, Gardette V, Julien S, Covas R, Gomez D, Doutrelant C (2019) The theory of island biogeography and soundscapes: species diversity and the organization of acoustic communities. J Biogeogr 46:1901–1911. https://doi.org/10.1111/jbi.13611
Robert A, Melo M, Lengagne T, Julien S, Gomez D, Doutrelant C (2021) Patterns of bird song evolution on islands support the character release hypothesis in tropical but not in temperate latitudes. J Evol Biol 34:1580–1591. https://doi.org/10.1111/jeb.13928
Robert A, Lengagne T, Melo M, Gomez D, Doutrelant C (2022) Evolution of vocal performance and song complexity in island birds. J Avian Biol 2022:e02726. https://doi.org/10.1111/jav.02726
Salerno PE, Chan LM, Pauly GB, Funk WC, Robertson JM (2022) Near-shore island lizard fauna shaped by a combination of human-mediated and natural dispersal. J Biogeogr 50:116–129. https://doi.org/10.1111/jbi.14511
Salvin O, Godman F (1892) Thamnophilus atrinucha. In: Biologia Centrali–Americana Aves, vol 2. Porter, London, p 200. https://doi.org/10.5962/bhl.title.54584
Sebastianelli M, Blumstein DT, Kirschel ANG (2021) Higher-pitched bird song towards the coast supports a role for selection in ocean noise avoidance. Bioacoustics 31:41–58. https://doi.org/10.1080/09524622.2021.1879680
Seddon N (2005) Ecological adaptation and species recognition drives vocal evolution in neotropical suboscine birds. Evolution 59:200–215
Seddon N, Tobias JA (2006) Duets defend mates in a sub- oscine passerine, the warbling antbird (Hypocnemis cantator). Behav Ecol 17:73–83. https://doi.org/10.1093/beheco/ari096
Sementili-Cardoso G, Gonçalves F, Martos R, Whitacker R, Marques R, Donatelli RJ (2018) Variation among vocalizations of Taraba major (Aves: Thamnophilidae) subspecies. Stud Neotrop Fauna E 53:120–131. https://doi.org/10.1080/01650521.2017.141498
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90
Shipilina D, Serbyn M, Ivanitskii V, Marova I, Backström N (2017) Patterns of genetic, phenotypic, and acoustic variation across a chiffchaff (Phylloscopus collybita abietinus/tristis) hybrid zone. Ecol Evol 7:2169–2180. https://doi.org/10.1002/ece3.2782
Slabbekoorn H, Smith TB (2002) Bird song, ecology and speciation. Phil Trans R Soc B 357:493–503. https://doi.org/10.1098/rstb.2001.1056
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4(1):vey016. https://doi.org/10.1093/ve/vey016
Swofford DL (2003) PAUP* Phylogenetic Analysis Using Parsimony, version 4.0a. Sinauer Associates, Sunderland
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–95 (PMC1203831)
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Tarwater CE (2012) Influence of phenotypic and social traits on dispersal in a family living, tropical bird. Behav Ecol 23:1242–1249. https://doi.org/10.1093/beheco/ars108
Tarwater CE, Brawn JD, Maddox JD (2013) Low extrapair paternity observed in a tropical bird despite ample opportunities for extrapair mating. Auk 130:733–741. https://doi.org/10.1525/auk.2013.13117
Thayer JE, Bangs O (1905) The vertebrata of Gorgona island, Colombia: Aves. Bull Mus Comp Zool 46:98–102
Theron E, Hawkins K, Bermingham E, Ricklefs RE, Mund NI (2001) The molecular basis of an avian plumage polymorphism in the wild. Curr Biol 11:550–557. https://doi.org/10.1016/s0960-9822(01)00158-0
Thom G, Aleixo A (2015) Cryptic speciation in the white-shouldered antshrike (Thamnophilus aethiops, Aves – Thamnophilidae): The tale of a transcontinental radiation across rivers in lowland Amazonia and the northeastern Atlantic Forest. Mol Phylogenet Evol 82:95–110. https://doi.org/10.1016/j.ympev.2014.09.023
Tobias JA, Aben J, Brumfield RT, Derryberry EP, Halfwerk W, Slabbekoorn H, Seddon N (2010) Song divergence by sensory drive in Amazonian birds. Evolution 64:2820–2839. https://doi.org/10.1111/j.1558-5646.2010.01067.x
Torres IMD, Barreiros MHM, de Araújo CB (2020) The acoustic ecology of an Amazonian bird assemblage: the role of allometry, competition and environmental filtering in the acoustic structure. Ibis 162:991–1000. https://doi.org/10.1111/ibi.12720
Touchton JM, Seddon N, Tobias JA (2014) Captive rearing experiments confirm song development without learning in a tracheophone suboscine bird. PLoS ONE 9:e95746. https://doi.org/10.1371/journal.pone.0095746
Valderrama E, Pérez-Emán JL, Brumfield RT, Cuervo AM, Cadena CD (2014) The influence of the complex topography and dynamic history of the montane Neotropics on the evolutionary differentiation of a cloud forest bird (Premnoplex brunnescens, Furnariidae). J Biogeogr 41:1533–1546. https://doi.org/10.1111/jbi.12317
Whitney BM, Oren DC, Brumfield RT (2004) A new species of Thamnophilus antshrike (Aves: Thamnophilidae) from the Serra Do Divisor, Acre, Brazil. Auk 121(1031):1039. https://doi.org/10.2307/4090473
Whittaker RJ, Fernandez-Palacios JM (2007) Island biogeography ecology, evolution and conservation. Oxford University Press, Oxford
Whittaker RJ, Fernández-Palacios JM, Matthews TJ, Borregaard MK, Triantis KA (2017) Island biogeography: taking the long view of nature’s laboratories. Science 357:eaam8326. https://doi.org/10.1126/science.aam8326
Wilkins MR, Seddon N, Safran RJ (2013) Evolutionary divergence in acoustic signals: causes and consequences. Trends Ecol Evol 28:156–166. https://doi.org/10.1016/j.tree.2012.10.002
Wilson AG, Chan Y, Taylor SS, Arcese P (2015) Genetic divergence of an avian endemic on the Californian Channel Islands. PLoS ONE 10:e0134471. https://doi.org/10.1371/journal.pone.0134471
Wolf LL (1977) Species relationships in the avian genus Aimophila. Ornithol Monogr 23:1–220. https://doi.org/10.2307/40166744
Zink RM, Barrowclough GF (2008) Mitochondrial DNA under siege in avian phylogeography. Mol Ecol 17(9):2107–2121. https://doi.org/10.1111/j.1365-294X.2008.03737.x
Zuccon A, Zuccon D (2014) MrEnt: an editor for publication-quality phylogenetic tree illustrations. Mol Ecol Resour 14:1090–1094. https://doi.org/10.1111/1755-0998.12253
Acknowledgements
We thank editor H. Brumm as well as C. Martin, G. Voelker, and four anonymous reviewers for dedicating their time and effort to review the manuscript. We sincerely appreciate their insightful comments and suggestions, which have significantly contributed to improving the manuscript.We appreciate the contributions of field assistants L. Calvert, H. Calero, and C. Guerrero in Gorgona Island and Guapi, as well as the support provided by H. Mantilla in Chocó and J. Calderon in Nariño. We extend our gratitude to the members of the Ecologia y Evolución de Vertebrados Research Group at Universidad de Antioquia, who offered valuable support in the field. Thanks to C. Muñoz as laboratory assistant and D. Gil for her assistance in formatting some sections of the manuscript. We thank Dr. M. Mosquera-Escudero at the Physiological Science Laboratory, Universidad del Valle. We also thank the Vice Rectories of Research of both Universidad del Valle and Universidad de Antioquia. Furthermore, we thank L. Payan and X. Zorrilla, the kind administrative staff of the Gorgona Island National Park. Specimens collected were placed in the ornithological collections at both Universidad del Valle and Universidad de Antioquia.
Funding
Open Access funding provided by Colombia Consortium This work was funded by Universidad del Valle, CO and Universidad de Antioquia, CO.
Author information
Authors and Affiliations
Contributions
RSC: conceptualization, methodology, data collection, formal analysis in phylogenetics and phylogeography, resources, writing-original draft, writing-review and editing, visualization, supervision, funding acquisition. NPA: fieldwork, data collection, data curation, writing-review. HRG: conceptualization, methodology, data collection, data curation in acoustics, formal analysis in acoustics, resources, writing-review and editing, visualization, funding acquisition.
Corresponding author
Ethics declarations
Ethics approval
Universidad del Valle and Universidad de Antioquia provided additional research permits and approval from ethical committees. Institutional guidelines for the use of animals were followed. Permits approval for collecting specimens in Colombia has been obtained for this study to characterized avian evolutionary significant units. These permits are up to date for RS by Universidad del Valle and to HRF by Universidad de Antioquia, Colombia. The capture/collection of biological samples and specimens is covered by permits as follows: (i) A permit for the collection of specimens for non-commercial purposes, issued by the National Environmental Licensing Authority (ANLA) through resolutions 05240 and 01053 to the Universidad de Antioquia and Universidad del Valle respectively, which includes the Ecology and Evolution research group and Animal Ecology research group on the latter permit. (ii) A permit for collecting specimens for non-commercial purposes within areas of the national park system, issued by the Ministry of Environment and Sustainable Development through resolution 0120 to the Universidad del Valle. Thanks to the Unidad Administrativa Especial del Sistema de Parques Nacionales Naturales de Colombia.
Competing interests
None.
Additional information
Communicated by H. Brumm
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sedano-Cruz, R.E., Pérez-Amaya, N. & Rivera-Gutierrez, H.F. Vocal and genetic variation between a land-bridge island and mainland populations of the Black-crowned Antshrike (Thamnophilus atrinucha). Behav Ecol Sociobiol 78, 42 (2024). https://doi.org/10.1007/s00265-024-03455-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00265-024-03455-6