Abstract
Purpose
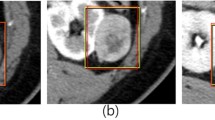
With advancements in medical imaging, more renal tumors are detected early, but it remains a challenge for radiologists to accurately distinguish subtypes of renal parenchymal tumors. We aimed to establish a novel deep convolutional neural network (CNN) model and investigate its effect on identifying subtypes of renal parenchymal tumors in T2-weighted fat saturation sequence magnetic resonance (MR) images.
Methods
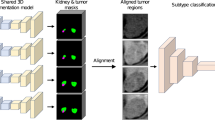
This retrospective study included 199 patients with pathologically confirmed renal parenchymal tumors, including 77, 46, 34, and 42 patients with clear cell renal cell carcinoma (ccRCC), chromophobe renal cell carcinoma (chRCC), angiomyolipoma (AML), and papillary renal cell carcinoma (pRCC), respectively. All enrolled patients underwent kidney MR scans with the field strength of 1.5 Tesla (T) or 3.0 T before surgery. We selected T2-weighted fat saturation sequence images of all patients and built a deep learning model to determine the type of renal tumors. Receiver operating characteristic (ROC) curve was depicted to estimate the performance of the CNN model; the accuracy, precision, sensitivity, specificity, F1-score, and area under the curve (AUC) were calculated. One-way analysis of variance and χ2 tests of independent samples were used to analyze the variables.
Results
The experimental results demonstrated that the model had a 60.4% overall accuracy, a 61.7% average accuracy, and a macro-average AUC of 0.82. The AUCs for ccRCC, chRCC, AML, and pRCC were 0.94, 0.78, 0.80, and 0.76, respectively.
Conclusion
Deep CNN model based on T2-weighted fat saturation sequence MR images was useful to classify the subtypes of renal parenchymal tumors with a relatively high diagnostic accuracy.





Similar content being viewed by others
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Kim Y, Sung D, Sim K, et al. (2017) Renal tumors with low signal intensities on T2-weighted MR image: radiologic-pathologic correlation. Abdom Radiol (NY) 42:2108-2118.
Farrell C, Noyes S, Tourojman M, Lane B. (2015) Renal angiomyolipoma: preoperative identification of atypical fat-poor AML. Curr Urol Rep 16:12.
Mehta V, Venkataraman G, Antic T, Rubinas TC, Le Pool IC, Picken MM. (2013) Renal angiomyolipoma, fat-poor variant – a clinicopathologic mimicker of malignancy. Virchows Arch 463:41-46.
Ljungberg B, Bensalah K, Canfield S, et al. (2015) EAU Guidelines on renal cell carcinoma: 2014 update. Eur Urol 67:913-924.
Tzortzakakis A, Gustafsson O, Karlsson M, Ekström-Ehn L, Ghaffarpour R, Axelsson R. (2017) Visual evaluation and differentiation of renal oncocytomas from renal cell carcinomas by means of 99mTc-sestamibi SPECT/CT. Ejnmmi Res 7:29.
Wang K, Sun Y, Tao W, Fei X, Chang C. (2017) Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett 394:1-12.
Schieda N, Pol CB, Moosavi B, McInnes MDF, Mai K, Flood T. (2015) Intracellular lipid in papillary renal cell carcinoma (pRCC): T2 weighted (T2W) MRI and pathologic correlation. Eur Radiol 25:2134-2142.
Keegan KA, Schupp CW, Chamie K, et al. (2012) Histopathology of surgically treated renal cell carcinoma: survival differences by subtype and stage. J Urol 188(2): 391-397.
Ficarra V, Martignoni G, Galfano A, Novara G, Gobbo S, Brunelli M, Pea M, Zattoni F, Artibani W. (2006) Prognostic role of the histologic subtypes of renal cell carcinoma after slide revision. Eur Urol 50(4): 786–794.
Klatte T, Han K, Said JW, et al. (2008) Pathobiology and prognosis of chromophobe renal cell carcinoma. Urol Oncol 26:604–609.
Lee S, Fuerst B, Tateno K, et al. (2017) Multi-modal imaging, model-based tracking, and mixed reality visualisation for orthopaedic surgery. Health Technol Lett 4:168-173.
Dong L, Zhang P, Lei P, et al. (2017) PEGylated GdF3: Fe Nanoparticles as multimodal T1/T2-weighted MRI and X-ray CT imaging contrast agents. ACS Appl Mater Interfaces 9:20426-20434.
Mazonakis M, Damilakis J. (2016) Computed tomography: what and how does it measure? Eur J Radiol 85:1499-1504.
Bruno F, Arrigoni F, Mariani S, et al. (2019) Advanced magnetic resonance imaging (MRI) of soft tissue tumors: techniques and applications. Radiol Med 124:243-252.
Degirmenci A, Perrin DP, Howe RD. (2018) High dynamic range ultrasound imaging. Int J Comput Assist Radiol Surg 13:721-729
Bibault JE, Giraud P, Burgun A. (2016) Big data and machine learning in radiation oncology: state of the art and future prospects. Cancer Letts 382:110-117.
van Oostenbrugge TJ, Fütterer JJ, Mulders, PF. (2018) Diagnostic Imaging for Solid Renal Tumors: A Pictorial Review. Kidney Cancer 2(2):79–93.
Hylton NM, Blume JD, Bernreuter WK. (2012) Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy – results from ACRIN 6657/I-SPY TRIAL. Radiology 263:663-672.
Zhou J, Li GJ, Sheng FG, Qiao PG. (2016) Magnetic resonance imaging evaluation of residual tumors in breast cancer after neoadjuvant chemotherapy: surgical implications. Acta Radiol 57:529-537.
Blachar A, Federle MP, Ferris JV, et al. (2018) Radiologists’ performance in the diagnosis of liver tumors with central scars by using specific CT criteria. Radiology 223:532-539.
Lecun Y, Bengio Y, Hinton G. (2015) Deep learning. Nature 521:436-444.
Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. (2018) Deep learning with convolutional neural network in radiology. Jpn J Radiol 36:257-272.
Acharya UR, Oh SL, Hagiwara Y, et al. (2017) A deep convolutional neural network model to classify heartbeats. Comput Biol Med 89:389-396.
Brinker TJ, Hekler A, Enk AH, et al. (2019) A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer 111:148-154.
Setio AAA, Traverso A, de Bel T, et al. (2017) Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 42:1-13.
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. (2018) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79:2379-2391.
Yasaka K, Akai H, Abe O, Kiryu S. (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887-896
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207-1216.
Tabibu S, Vinod PK, Jawahar CV. (2019) Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep 9(1):1-9.
Han S, Hwang SI, Lee HJ. (2019) The classification of renal cancer in 3-phase CT images using a deep learning method. J Digit Imaging 32(4):638-643.
He K, Zhang X, Ren S, et al. (2016) Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.
Feng Z, Rong P, Cao P, et al. (2018) Machine learning-based quantitative texture analysis of CT images of small renal masses: differentiation of angiomyolipoma without visible fat from renal cell carcinoma. Eur Radiol 28:1625-1633.
Kocak B, Yardimci AH, Bektas CT, et al. (2018) Textural differences between renal cell carcinoma subtypes: machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur J Radiol 107:149-157.
Lee H, Hong H, Kim J, Jung DC. (2018) Deep feature classification of angiomyolipoma without visible fat and renal cell carcinoma in abdominal contrast-enhanced CT images with texture image patches and hand-crafted feature concatenation. Med Phys 45:1550-1561.
Korotcov A, Tkachenko V, Russo DP, Ekins S. (2017) Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol Pharm 14:4462-4475.
Shen D, Wu G, Suk HI. (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221-248.
Zhou L, Zhang Z, Chen YC, Zhao ZY, Yin XD, Jiang HB. (2019) A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Transl Oncol 12:292-300.
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary phase MR images. Radiology 287:146-155.
Xi IL, Zhao Y, Wang R, et al. (2020) Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin Cancer Res 26(8):1944–1952.
Fenstermaker M, Tomlins SA, Singh K, et al. (2020) Development and Validation of a Deep-learning Model to Assist With Renal Cell Carcinoma Histopathologic Interpretation. Urology 144:152–157.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional and/or National Research Committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, Y., Wang, S., Chen, Y. et al. Deep learning with a convolutional neural network model to differentiate renal parenchymal tumors: a preliminary study. Abdom Radiol 46, 3260–3268 (2021). https://doi.org/10.1007/s00261-021-02981-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-021-02981-5