Abstract
Purpose
In radioguided surgery (RGS), radiopharmaceuticals are used to generate preoperative roadmaps (e.g., PET/CT) and to facilitate intraoperative tracing of tracer avid lesions. Within RGS, there is a push toward the use of receptor-targeted radiopharmaceuticals, a trend that also has to align with the surgical move toward minimal invasive robotic surgery. Building on our initial ex vivo evaluation, this study investigates the clinical translation of a DROP-IN β probe in robotic PSMA-guided prostate cancer surgery.
Methods
A clinical-grade DROP-IN β probe was developed to support the detection of PET radioisotopes (e.g., 68 Ga). The prototype was evaluated in 7 primary prostate cancer patients, having at least 1 lymph node metastases visible on PSMA-PET. Patients were scheduled for radical prostatectomy combined with extended pelvic lymph node dissection. At the beginning of surgery, patients were injected with 1.1 MBq/kg of [68Ga]Ga-PSMA. The β probe was used to trace PSMA-expressing lymph nodes in vivo. To support intraoperative decision-making, a statistical software algorithm was defined and optimized on this dataset to help the surgeon discriminate between probe signals coming from tumors and healthy tissue.
Results
The DROP-IN β probe helped provide the surgeon with autonomous and highly maneuverable tracer detection. A total of 66 samples (i.e., lymph node specimens) were analyzed in vivo, of which 31 (47%) were found to be malignant. After optimization of the signal cutoff algorithm, we found a probe detection rate of 78% of the PSMA-PET-positive samples, a sensitivity of 76%, and a specificity of 93%, as compared to pathologic evaluation.
Conclusion
This study shows the first-in-human use of a DROP-IN β probe, supporting the integration of β radio guidance and robotic surgery. The achieved competitive sensitivity and specificity help open the world of robotic RGS to a whole new range of radiopharmaceuticals.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Being one of the most widely applied forms of image-guided surgery, radioguided surgery (RGS) is an interventional nuclear medicine technique that directs surgeons toward tissue targets preoperatively defined on imaging roadmaps such as PET/CT and SPECT/CT [1, 2]. To this aim, following initial diagnostics, a radiopharmaceutical is injected into the patient before surgery, and the surgeon is given an intraoperative detector enabling the real-time identification of areas with accumulated radiotracer.
Today, most RGS applications are based on the detection of low-energy (< 150 keV) γ-emitting radiopharmaceuticals (e.g., 99mTc-based tracers). Localization of 99mTc avid lesions is performed using intraoperative γ probes that provide numerical and acoustical feedback proportional to the counting rate and, thus, to the amount of radiopharmaceutical detected in the considered sample. A unique characteristic of the γ-based RGS approach is the long penetration of such radiation in human tissue. In fact, if we consider 99mTc, 1/3 of its photons penetrate more than 8 cm of tissue. This property holds both advantages and disadvantages. It allows for the detection of “deeply located” lesions, but shine-through can pose a problem when the target tissue is located in an area characterized by elevated physiological uptake. Originally RGS was pursued in open surgery, but in recent years, detector technologies have adapted to support even robot-assisted surgery, e.g., via the DROP-IN γ probe [3, 4].
Since its introduction in the form of radioimmunoguided surgery (RIGS), tumor-targeted RGS is today undergoing a resurgence, a prime example being the introduction of PSMA-targeted surgery in prostate cancer patients [5]. Due to relatively low uptake values and more complex tracer pharmacokinetics, these receptor-targeted procedures have proven to be more challenging to perform than, e.g., sentinel lymph node (SLN) procedures [6]. Comparative analysis showed that signal-to-background ratios (SBRs) influence surgical decision-making in the context of RGS.
Given the focus of global radiochemistry efforts on the production of receptor-specific PET tracers, it makes sense to investigate if and how tracers already used in patients for PET diagnostics can also find use in RGS applications, with the aim of providing a complementary approach with respect to SLN procedures [6].
In this context, we have exploited the use of β particle detectors [7, 8]. Since β radiation undergoes more tissue attenuation, penetration is reduced to a few mms, and this feature could help mitigate the shine-through phenomenon [9]. The impact of this effect on SBR values builds on previous studies with the PET radiotracer [68Ga]Ga-PSMA using Monte Carlo simulations [10] and initial ex vivo evaluations [11]. In general, RGS is based on giving the surgeon real-time (numeric/acoustic) feedback of the amount of tracer that is accumulated in the area being sampled with the probe. However, especially for receptor-targeted procedures, only in very particular cases, this feedback is so unambiguous enough to allow immediate discrimination of tumors from healthy tissue. Indeed PET images as well as common clinical experience suggest that tissue uptake may vary significantly, not only between tumor and healthy tissue, but also among lesions. Discussion also remains on what should be defined as “background” during surgery [12]. Finally, there is also no agreement yet on what “a fixed, arbitrary” SBR cutoff should be defined to consider a tissue sample “positive” or “negative”; SBRs of 2 [13, 14] and 1.5 [15] were previously suggested for image guidance.
In the current study, we clinically translated a prototype DROP-IN β probe to investigate, for the first time, its in vivo use in prostate cancer patients. In particular, we studied how the technology can support the intraoperative decision-making process. Utilizing typical ROC curve analysis, we concentrated on the development of a statistically sound signal-to-background discrimination algorithm able to optimize the sensitivity and specificity of the tracing technique. This innovative approach was compared to the standard “fixed SBR method” that uses the cutoff values 1.5 and 2.
Material and methods
Engineering a clinical-grade DROP-IN β probe
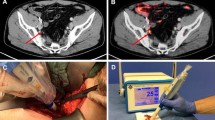
The sterilizable DROP-IN set-up (see Fig. 1) consists of four main parts: (1) a β particle detector, (2) a DROP-IN housing, (3) an electronic processing and read-out unit, and (4) a dedicated statistical software algorithm for signal interpretation.
The core of the β detector itself consists of a cylindrical (6 mm diameter and 3 mm height) mono-crystalline para-terphenyl (doped with 0.1% in mass of (E,E)-1,4-diphenyl-1,3- butadiene) [16]. This material has been shown to be highly effective for β + detection, as it is transparent to the 511 keV γ particles. Tests with 68 Ga source suggest that it has an ~ 80% efficiency to positrons above ~ 110 keV while being, at the same time, substantially transparent to photons of ~ 500 keV, with an efficiency of ~ 3% [7, 17]. This intrinsic γ transparency therefore limits the need for collimation. The scintillation crystal is surrounded by a 1-mm-thick external black polyvinyl chloride (PVC) ring, combined with the housing described below.
Scintillation light conversion was performed by a 3 × 3 mm2 silicon photomultiplier (SiPM Hamamatsu S13360-3050PE) powered and read-out by a custom microcontroller, connected to the device with a biocompatible and sterilizable latex-free cable.
The DROP-IN housing hosting the crystal and the SiPM was designed to be picked up with the robotic steerable instruments (e.g., ProGrasp Forceps, Intuitive). The DROP-IN shape and diameter (11 mm) were sized to make it compatible with typical trocar dimensions (maximum 12–15 mm). Length (45 mm) and weight (6 g) were optimized to also facilitate maneuverability.
The housing was realized in PEEK (polyetheretherketone): a light, water-tight, sterilizable, robust, and biocompatible plastic material. To meet the required light tightness, black PEEK was chosen. The housing lateral wall thickness was 1.5 mm, while β-detection sensitivity at the front of the detector was maximized by machining the PEEK wall to be as thin as 300 µm.
The electronics then translate the analog signal produced by β particle crossing into both numerical (i.e., counts/s) and audible feedback. Exposure time (or counting time) was chosen as 1 s.
Since the definition of the most optimal statistical algorithm to guide surgical decision-making was, as already described, one of the aims of this study, no further visual information was shown to the surgeon during the operation, apart from a strip chart of the acquired countings (see Fig. 1).
Patients’ population and surgical procedure
The clinical study was performed at IEO Milano (Institutional Review Board of the European Institute of Oncology, protocol code UID 1703, clincialtrial.gov ID NCT05596851) and considered 7 primary prostate cancer patients, scheduled for radical prostatectomy combined with extended pelvic lymph node dissection. All patients had high-risk prostate cancer according to EAU classification, a tumor stage of cT3 and Gleason scores between + 3 and 4 + 5, and a positive diagnostic PSMA-PET/CT scan with at least 1 lymph node metastasis visible. To enable β RGS, all patients received a second low-dose injection of [68Ga]Ga-PSMA-11 (about 1.1 MBq/kg) directly in the operating room prior to surgery.
The surgical resection was performed with a da Vinci Xi robotic surgical platform, and lymph node dissection preceded the prostatectomy. Beta probe scanning/tracing was performed, following the template, on a district basis. A district is defined as an area sharing the same definition of tracer background. For each district scanned, the surgeon was instructed to follow this procedure:
-
1.
Perform a background acquisition in the given district. The probe was used to measure background for at least 5–10 s, keeping it in contact with the tissue, in an area assumed free of disease.
-
2.
Use the probe to analyze different lymph nodes in the considered district according to the followed surgical template, with the possibility to perform more than one measurement over the same sample.
The surgeon was left free to decide at their judgment how many times and where to perform background acquisitions, for example, when moving to another, distant zone or if a significant time had elapsed.
To easily compare measurements acquired at different times, all probe counts were corrected for the physical decay of 68 Ga. Only lymph nodes were analyzed with the probe, and all in vivo measurements were recorded for offline investigation (raw data + da Vinci video + probe video). To ensure no positive lesions were missed, every excised sample was checked ex vivo with a second β probe based on the same technology used for the DROP-IN prior to sending it out for standard pathological examination.
Signal-to-background ratio discrimination algorithm
In a previous ex vivo study on GEPNET samples, using [90Y]Y-DOTATOC, a statistical approach for the discrimination algorithm was proposed and tested, leading to a sensitivity of 93% and specificity of 100% [18, 19]. Here, the cutoff was defined as follows:
where \(<{{R}_{Bkg}}^{{\text{Flat}}}>=({{R}^{{\text{Bkg}}}}_{{\text{Max}}}+{{R}^{{\text{Bkg}}}}_{{\text{Min}}})/2\) is the expected value of background countings, assuming them to have a flat distribution, \({\sigma }_{{\text{Flat}} }=({{R}_{{\text{Bkg}}}}^{{\text{Max}}}-{{R}_{{\text{Bkg}}}}^{{\text{Min}}})/\sqrt{12}\) is the corresponding variance, and \({N}_{\sigma }\) is chosen to be 3.
In the higher background context faced by β+-RGS, using [68Ga]Ga-PSMA, such a discrimination algorithm becomes more complex, as the challenge is to identify efficiently tumor samples while avoiding accidental background signals of 511 keV γ rays.
In this study, we set out to identify and optimize the best way to adapt this cutoff approach, considering two possible ways a positive lesion can present.
A sample is rated “Probe-Positive” if either one of these two conditions is satisfied:
where \({N}_{{\text{AboveCutoff}}}\) is the number of countings above a threshold \({R}_{{\text{Cutoff}}}\), \({N}_{{\text{Tot}}}\) is the total number of countings in the sample, and \({\epsilon }_{{\text{Frac}}}\) is a parameter of the algorithm to be defined or
where \({N}_{{\text{AboveCutoff}}}^{{\text{Contiguous}}}\) is the length of the longest series of contiguous countings exceeding \({R}_{{\text{Cutoff}}}\) and \({\epsilon }_{{\text{Num}}}\) is another parameter of the algorithm.
In order to fully exploit the discriminating power of the probe, \({R}_{{\text{Cutoff}}}\) is defined as in Eq. 1 but leaving \({N}_{\sigma }\) free to vary.
How these two conditions allow to identify possible lesions having different countings topologies is clarified in Fig. 2.
Effect of the two detection conditions (Eqs. 2 and 3). In this example, a 20 s acquisition was performed on a sample, having a cutoff value of 100 CPS (horizontal line), considering the algorithm parameters: \({\upepsilon }_{{\text{Num}}}=4\) and \({\upepsilon }_{{\text{Frac}}}=30\mathrm{\%}\). The color of the legend text suggests whether the two detection conditions were satisfied (red, indicating tumor) or not (green, indicating free of tumor). A red outline of the total legend means that the algorithm identified the sample as Probe + (indicating tumor)
To find the best possible settings of this approach, thus delivering the best sensitivity and specificity as compared to pathological results, the combination of these 3 parameters was optimized:
-
The number of sigmas above the mean value we set the cutoff to, \({N}_{\sigma }\) (see Eq. 1) that we varied to assume the values {0.5, 1, 1.5, 2, 2.25, 2.5, 2.75, 3, 3.25, 3.5, 3.75, 4, 4.5, 5, 6, 8, 10, 30} (\({{N}^{{\text{Values}}}}_{\sigma }=18\))
-
The minimum fraction of countings above a signal in a sample \({\epsilon }_{{\text{Frac}}}\) (see Eq. 2) that we varied in the ensemble {5%, 10%, 20%, 50%} (\({{N}^{{\text{Values}}}}_{{\epsilon }_{{\text{Frac}}} }=4)\)
-
The minimum number of consecutive countings above a threshold \({\epsilon }_{{\text{Num}}}\) (see Eq. 3) that we allowed to assume the values {2, 3, 4, 6, 8, 10, 20} (\({{N}^{{\text{Values}}}}_{{\epsilon }_{{\text{Num}}} }=7)\)
All things considered, for each sample, the same data were analyzed with a combination of 504 (\({{N}^{{\text{Values}}}}_{\sigma }\times {{N}^{{\text{Values}}}}_{{\epsilon }_{{\text{Frac}}} }\times {{N}^{{\text{Values}}}}_{{\epsilon }_{{\text{Num}}}}\)) different parameters, for each of which the sample could be defined as “Probe-Positive” or “Probe-Negative.” This result was then compared with pathological analysis, considered as the standard reference, defining each sample as Path.-Positive or Path.-Negative and thus allowing to classify it as true positive (TP), true negative (TN), false positive (FP), or false negative (FN).
Therefore, for each chosen value Nσ, sensitivity and specificity can be calculated as.
And, for a given pair of ϵFrac and ϵNum, a ROC curve can be constructed by plotting the variation of sensitivity and specificity with Nσ.
To strengthen the analysis, the whole dataset was split into two parts: one to be used for training the algorithm (training dataset) and the other to test it (test dataset). To this aim, data were randomly split on a district basis and assigned to either the training or test dataset, in order to find a distribution of districts between the two samples that contain, according to pathology, a similar number of healthy and diseased areas.
Results
DROP-IN probe: clinical translation and data collection
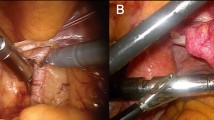
Before every procedure, the medical grade DROP-IN β probe was sterilized with a standard gas-plasma sterilization cycle. During the surgical procedures, it was used through a standard 12-mm assistant trocar (see Fig. 3). Pick-up of the DROP-IN β probe was simple using the standard da Vinci tools (e.g., ProGrasp Forceps instrument). The surgeon was able to use the probe to scan the field autonomously exploiting all 6 available degrees of freedom, without the need of any help from the assistant. In each patient, the DROP-IN probe was used to perform lymph node examination for around 20 min in total, having therefore reduced impact on surgery duration. No complications related to the use of the prototype probe nor of the [68Ga]Ga-PSMA tracer were observed, and no issues were encountered in the sterilization procedure.
A Operating room during one of the procedures: the black cable coming from the drop-in probe can be distinguished. B MIP image of 68 Ga-PSMA-PET of one of the considered patients (Pt. #3). C View of the da Vinci monitor as seen by the surgeon during the procedure (Pt. #5), including the TilePro split-screen option to view the probe measurements directly in the surgical console
Seven patients were considered in this study, and surgery was performed between Jan 2023 and Oct 2023. Median injected activity was 100 MBq (IQR 84–107). Probe measurements were performed in a time after injection ranging from 20 to 152 min (median 49 min, IQR 34–94). A total of 66 samples were analyzed with the probe in vivo, belonging to 25 districts (i.e., a group of samples for which the surgeon retained to use the same background evaluation). Of these 66 samples, pathology examination identified 31 to be bearing tumors (47% “Path.-Positive”) and 35 not (“Path.-Negative”), while only 19 were PET-positive and 47 PET-negative. Patients’ data are summarized in Table 1.
Signal-to-background cutoff value: identification and testing
As described in the “Signal-to-background ratio discrimination algorithm” section, the surgically collected dataset was used to statistically calculate the best SBR discrimination algorithm for surgery, i.e., the one having the most optimal sensitivity and specificity as compared to pathology. To this end, the dataset was randomly split in such a way as to have two sub-datasets comprising a similar amount of positive and negative samples. In particular, among all possible combinations, the train dataset contained a total of 35 samples, while the test dataset contained 31 samples. The procedure described in the “Signal-to-background ratio discrimination algorithm” section was then applied to the train dataset: 28 (= \({{N}^{{\text{Values}}}}_{{\upepsilon }_{{\text{Frac}}} }\times {{N}^{{\text{Values}}}}_{{\epsilon }_{{\text{Num}}}}\)). ROC curves, having 18 (= \({{N}^{{\text{Values}}}}_{\sigma }\)) points each, were constructed (see Fig. 4A).
A ROC curves obtained as described in the “Signal-to-background ratio discrimination algorithm” section for the train dataset. The circled point represents the most effective cutoff, corresponding to \({\upepsilon }_{{\text{Num}}}=10\), \({\upepsilon }_{{\text{Frac}}}=5\mathrm{\%}\), and \({N}_{\upsigma }=4.5\). B ROC curve obtained for the test dataset, in the case identified as the most effective cutoff in the train dataset (whose sensitivity and specificity are also shown for reference in the red circle). The yellow-circled point represents the performances of this same cutoff in the test dataset. Numbers nearby points refer to the nsigma of the given point
All ROCs were found to have a high area under the curve (AUC) (median 0.911, IQR 0.905–0.921). The best point in this curve, representing the most effective cutoff on this train dataset, is circled in Fig. 4A and was chosen as the one nearest to the 100 − 100 reference value. This was found to be the point relative to \({\epsilon }_{{\text{Num}}}=10\), \({\epsilon }_{{\text{Frac}}}=5\%\), and \({N}_{\sigma }=4.5\), having sensitivity = 86% and specificity = 90%.
Figure 4B compares this best-performing point in the train dataset (red circle) with the corresponding value in the test dataset (yellow circle), found to have 76–93% sensitivity–specificity. Considering the relatively small amount of data points available for this analysis after the splitting, similar values for sensitivity and specificity were therefore found between the two datasets, thus confirming the robustness of the identified cutoff and more generally of the defined approach.
Application of the best cutoff value algorithm
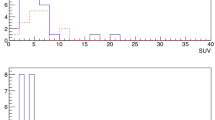
To demonstrate how the suggested signal-to-background cutoff algorithm could assist surgical decision-making in future in vivo studies, Fig. 5 visualizes a typical acquisition of a district in the current surgical dataset, using the best algorithm parameters. In the plot, blue data represent the background measurement phase, while orange and green data represent probe measurements performed respectively on Path.-Positive and Path.-Negative samples. In this specific district example of patient #2, the cutoff value (red horizontal dashed line) was found to be RCutoff = 153 CPS. All countings exceeding this cutoff are red dashed, but only samples also complying with “condition formula 2” or “condition formula 3”, with \({\epsilon }_{{\text{Num}}}=10\), \({\epsilon }_{{\text{Frac}}}=5\%\), are considered probe-positive and have therefore a red border around their label. In the particular case represented in Fig. 5, therefore, thanks to the discrimination algorithm, the probe would have identified 1 true positive and 2 true negatives.
A Example of data acquired when scanning a given district with the best discrimination algorithm. Blue data represent counts considered to evaluate the background, while orange and green ones represent measurements performed on tumor and healthy tissue respectively, according to pathology examination. The label reports the name and pathology code of the considered sample, while the color of its surrounding border represents whether the sample was identified as probe-positive (red) or probe-negative (green) by the algorithm. B, C Screenshots of the DROP-IN probe performing measurements on samples M and N respectively
Applying the best cutoff algorithm to the entire patient data obtained in this study provides the following clinical outcome: the probe was able to detect 15 out of 19 PSMA-PET-positive lesions (78% detection rate). However, from the 4 missed, two turned out to be PET false positives at pathology, increasing the probe detection rate to 88% compared to PET, while the remaining two were most probably missed due to the presence of healthy tissue between them and the probe in the surgical setting. Interestingly, the probe did find 10 true positive samples, based on pathology, that were not found on preoperative PET. Even if a detailed correlation of probe counting with sample dimension was outside the scope of this first study, and therefore, not all samples’ footprints were measured; the ones being PET − but Probe + were found to have a diameter of 3–4 mm. When taking pathology as the true standard, the probe sensitivity and specificity were found to be 81% and 91% respectively considering the whole dataset, values that are as expected in line with those found in train and test datasets.
To compare the performances of the investigated discrimination algorithm with the typical “fixed SBR cutoff approach,” sensitivity and specificity were also calculated for a fixed 1.5 SBR cutoff ratio (60 and 90%, respectively) and 2 SBR cutoff ratio (45 and 97%, respectively; see Fig. 6).
Discussion
We investigated the first-in-human translation of a medical grade DROP-IN β probe, a technology that supports the detection of β + emitting tracers, so-called “PET tracers” [20], such as [68Ga]Ga-PSMA-11, thereby extending the impact of the respective PSMA-PET imaging roadmaps on surgical interventions. A recent EANM position paper indicates that technical advancement is needed to fully exploit beta-RGS [21], and a Delphi study on PSMA-targeted surgery indicates that more education on this topic is desirable [22]. These are critical points that we address in our report.
In PSMA cancer management PSMA-targeted RGS provides a complementary value to SLN-RGS procedures, allowing targeting of both macro- and micro-metastases [6], concepts that rely heavily on the DROP-IN design. Similar to what was previously reported for the DROP-IN γ probes [23], the DROP-IN β probe helped to fully exploit the degrees of freedom of steerable robotic instruments. Since background and target signal intensities affect intraoperative decision-making and surgical performance [24], we used our first-in-human data to define and optimize a statistical software algorithm providing a quantified signal cutoff value that could support surgical decision-making. While a fixed SBR value is often utilized for this application [12], this approach lacks a real statistical foundation and quantified and sound reasoning as to which SBR value should be chosen in radioguided surgery applications. After optimization, the proposed statistical algorithm for the identification of probe-positive lymph nodes outperformed the use of SBR cutoffs of either 1.5 or 2, when considering both sensitivity and specificity. A limitation of the current study is the limited number of samples on which the algorithm was trained and applied, in particular, due to the need to split the dataset for training and testing purposes. The reproducibility of our results between the two independent sub-samples suggests that the high sensitivity and specificity values are not incidental. Ongoing studies focus on the optimization of the proposed cutoff algorithm to the clinical settings using a larger patient cohort and thus a larger amount of positive and non-positive lymph nodes. It could be investigated if the algorithm is transferable to γ-RGS as well or if it should be redefined for every procedure (i.e., a combination of probe, tracer, and surgical application). Lastly, further optimization of the technique will allow to tailor the amount of injected activity to the given case, thus reducing it further with respect to the current “half PET” starting value.
It needs to be highlighted that demonstrating clinical benefit was not in the scope of this initial first-in-human study. However, a recent systematic review on PSMA-targeted surgery [12] indicates that different groups are pursuing different strategies to surgically identify PSMA-positive lesions. To assess the impact of the technology presented in this study, it is critical to compare the sensitivity and specificity to other studies that investigated PSMA-targeted surgery in the primary setting [13, 25,26,27,28]. Table 2 indicates that despite the ~ 3-mm-range tissue penetration of the DROP-IN β radiation, its sensitivity for lymph node detection seems to be at least as good as optimized near-infrared fluorescence guided surgery approaches which are assumed to support a larger signal penetration of < 10 mm. This is a rather unique finding, especially when one considers that the β-guidance approach requires about 300 times less of a PSMA-targeting tracer and that this can be the exact same tracer already used in the clinic for PSMA-PET. Combined, these features would help reduce costs and stimulate the dissemination of the image guidance concepts to hospitals that have access to [68Ga]Ga-PSMA-PET. The performance with β-guidance even equaled some of the γ-radioguidance examples, a finding that indicates that limiting the shine-through of background signals with beta detectors could provide a competitive edge.
The current study opens up a whole world of possible minimal invasive RGS applications [20]. Firstly, [68Ga]Ga-PSMA-guided surgery can be further explored, for example, also in the salvage setting for prostate cancer. There are then nevertheless also other possible options, like FDG-guided surgery (e.g., lung cancer [30]), carbonic anhydrase iX guided surgery (e.g., renal cell carcinoma [31]), C-MET guided surgery (e.g., renal cell carcinoma [32]), folate guided surgery (e.g., lung cancer [33]), and fibroblast-activation-protein guided surgery (i.e., > 28 different cancer types [34]).
To widen the technological impact, the use of 18F-based RGS is being investigated in parallel. Hereby, the results from a study that uses [18F]F-FDG in recurrent cervical cancer are encouraging [35]. Applications are expected to improve following technical refinements such as the exploitation of solid-state detectors that are more effective in combination with low-energy positrons as emitted by 18F, with respect to [68Ga] [36,37,38].
Conclusion
We show for the first time the clinical translation of a DROP-IN β probe that supports RGS in a robotic surgery setting. The obtained sensitivity and specificity values for nodal metastases were found to be competitive to values obtained for other PSMA-targeted surgery strategies.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Povoski SP, Neff RL, Mojzisik CM, O’Malley DM, Hinkle GH, Hall NC, et al. A comprehensive overview of radioguided surgery using gamma detection probe technology. World J Surg Oncol. 2009;7:11–11.
Van Oosterom MN, Rietbergen DDD, Welling MM, Van Der Poel HG, Maurer T, Van Leeuwen FWB. Recent advances in nuclear and hybrid detection modalities for image-guided surgery. Expert Rev Med Devices. 2019;16:711–34.
van Oosterom MN, Azargoshasb S, Slof LJ, van Leeuwen FWB. Robotic radioguided surgery: toward full integration of radio- and hybrid-detection modalities. Clin Transl Imaging. 2023. https://doi.org/10.1007/s40336-023-00560-w.
van Oosterom MN, Simon H, Mengus L, Welling MM, van der Poel HG, van den Berg NS, et al. Revolutionizing (robot-assisted) laparoscopic gamma tracing using a drop-in gamma probe technology. Am J Nucl Med Mol Imaging. 2016;6:1–17.
Horn T, Krönke M, Rauscher I, Haller B, Robu S, Wester HJ, et al. Single lesion on prostate-specific membrane antigen-ligand positron emission tomography and low prostate-specific antigen are prognostic factors for a favorable biochemical response to prostate-specific membrane antigen-targeted radioguided surgery in recurrent prostate cancer. Eur Urol. 2019;76:517–23.
van Leeuwen FWB, Winter A, van Der Poel HG, Eiber M, Suardi N, Graefen M, et al. Technologies for image-guided surgery for managing lymphatic metastases in prostate cancer. Nat Rev Urol. 2019;16:159–71.
Collamati F, Moretti R, Alunni-Solestizi L, Bocci V, Cartoni A, Collarino A, et al. Characterisation of a β detector on positron emitters for medical applications. Physica Med. 2019;67:85–90.
Camillocci ES, Baroni G, Bellini F, Bocci V, Collamati F, Cremonesi M, et al. A novel radioguided surgery technique exploiting β− decays. Sci Rep. 2015;4:4401–4401.
Heuveling DA, van Schie A, Vugts DJ, Hendrikse NH, Yaqub M, Hoekstra OS, et al. Pilot study on the feasibility of PET/CT lymphoscintigraphy with 89Zr-nanocolloidal albumin for sentinel node identification in oral cancer patients. J Nucl Med. 2013;54:585–9.
Collamati F, Bocci V, Castellucci P, De Simoni M, Fanti S, Faccini R, et al. Radioguided surgery with β radiation: a novel application with Ga68. Sci Rep. 2018;8.
Collamati F, van Oosterom MN, De Simoni M, Faccini R, Fischetti M, Mancini Terracciano C, et al. A DROP-IN beta probe for robot-assisted 68Ga-PSMA radioguided surgery: first ex vivo technology evaluation using prostate cancer specimens. EJNMMI Res. 2020;10(1):92. https://doi.org/10.1186/s13550-020-00682-6.
Berrens A-C, Knipper S, Marra G, van Leeuwen PJ, van der Mierden S, Donswijk ML, et al. State of the art in prostate-specific membrane antigen–targeted surgery—a systematic review. Eur Urol Open Sci. 2023;54:43–55.
Gandaglia G, Mazzone E, Stabile A, Pellegrino A, Cucchiara V, Barletta F, et al. Prostate-specific membrane antigen radioguided surgery to detect nodal metastases in primary prostate cancer patients undergoing robot-assisted radical prostatectomy and extended pelvic lymph node dissection: results of a planned interim analysis of a prospective phase 2 study. Eur Urol. 2022;82:411–8.
de Barros HA, van Oosterom MN, Donswijk ML, Hendrikx JJMA, Vis AN, Maurer T, et al. Robot-assisted prostate-specific membrane antigen–radioguided salvage surgery in recurrent prostate cancer using a DROP-IN gamma probe: the first prospective feasibility study. Eur Urol. 2022;82:97–105.
Azargoshasb S, Boekestijn I, Roestenberg M, KleinJan GH, van der Hage JA, van der Poel HG, et al. Quantifying the impact of signal-to-background ratios on surgical discrimination of fluorescent lesions. Mol Imaging Biol. 2023;25:180–9.
Angelone M, Battistoni G, Bellini F, Bocci V, Collamati F, De Lucia E, et al. Properties of para-terphenyl as a detector for α, β and γ radiation. IEEE Trans Nucl Sci. 2014;61:1483–7.
Mirabelli R, Morganti S, Cartoni A, De Simoni M, Faccini R, Fischetti M, et al. Characterization and optimization of a β detector for 18F radio-guided surgery. Phys Med. 2023;108:102545
Morganti S, Bertani E, Bocci V, Colandrea M, Collamati F, Cremonesi M, et al. Tumor-non-tumor discrimination by a β- detector for radio guided surgery on ex-vivo neuroendocrine tumors samples. Phys Med. 2020;72:96–102.
Bertani E, Collamati F, Colandrea M, Faccini R, Fazio N, Ferrari ME, et al. First ex vivo results of beta-radioguided surgery in small intestine neuroendocrine tumors with Y-90-DOTATOC. Cancer Biother Radiopharm. 2021;cbr.2020.4487-cbr.2020.4487.
Collamati F, van Oosterom MN, Hadaschik BA, Fragoso Costa P, Darr C. Beta radioguided surgery: towards routine implementation? Q J Nucl Med Mol Imaging. 2021;65:229–43.
Fragoso Costa P, Shi K, Holm S, et al. Surgical radioguidance with beta-emitting radionuclides; challenges and possibilities: a position paper by the EANM. Eur J Nucl Med Mol Imaging. 2024. https://doi.org/10.1007/s00259-023-06560-2.
Berrens A-C, Scheltema M, Maurer T, Hermann K, Hamdy FC, Knipper S, et al. Delphi consensus project on prostate-specific membrane antigen (PSMA)–targeted surgery—outcomes from an international multidisciplinary panel. Eur J Nucl Med Mol Imaging. 2023 Available from: https://doi.org/10.1007/s00259-023-06524-6.
Dell’Oglio P, Meershoek P, Maurer T, Wit EMK, van Leeuwen PJ, van der Poel HG, et al. A DROP-IN gamma probe for robot-assisted radioguided surgery of lymph nodes during radical prostatectomy. Eur Urol. 2021;79:124–32.
Azargoshasb S, de Barros HA, Rietbergen DDD, Dell’Oglio P, van Leeuwen PJ, Wagner C, et al. Artificial intelligence-supported video analysis as a means to assess the impact of DROP-IN image guidance on robotic surgeons: radioguided sentinel lymph node versus PSMA-targeted prostate cancer surgery. Adv Intell Syst. 2023;5:2300192.
Yılmaz B, Şahin S, Ergül N, Çolakoğlu Y, Baytekin HF, Sökmen D, et al. 99mTc-PSMA targeted robot-assisted radioguided surgery during radical prostatectomy and extended lymph node dissection of prostate cancer patients. Ann Nucl Med. 2022;36:597–609.
Gondoputro W, Scheltema MJ, Blazevski A, Doan P, Thompson JE, Amin A, et al. Robot-assisted prostate-specific membrane antigen–radioguided surgery in primary diagnosed prostate cancer. J Nucl Med. 2022;63:1659–64.
Nguyen HG, Berg NS van den, Antaris AL, Xue L, Greenberg S, Rosenthal JW, et al. First-in-human evaluation of a prostate-specific membrane antigen–targeted near-infrared fluorescent small molecule for fluorescence-based identification of prostate cancer in patients with high-risk prostate cancer undergoing robotic-assisted prostatectomy. Eur Urol Oncol. 2023. https://doi.org/10.1016/j.euo.2023.07.004.
Stibbe JA, de Barros HA, Linders DGJ, Bhairosingh SS, Bekers EM, van Leeuwen PJ, et al. First-in-patient study of OTL78 for intraoperative fluorescence imaging of prostate-specific membrane antigen-positive prostate cancer: a single-arm, phase 2a, feasibility trial. Lancet Oncol. 2023;24:457–67.
Siebinga H, oldeHeuvel J, Rijkhorst E-J, Hendrikx JJMA, De Wit- van der Veen BJ. The impact of peptide amount on tumor uptake to assess PSMA receptor saturation on 68Ga-PSMA-11 PET/CT in patients with primary prostate cancer. J Nucl Med. 2023;64:63–8.
Boekestijn I, Azargoshasb S, Schilling C, Navab N, Rietbergen D, van Oosterom MN. PET- and SPECT-based navigation strategies to advance procedural accuracy in interventional radiology and image-guided surgery. Q J Nucl Med Mol Imaging. 2021;65:244–60.
Povoski SP, Hall NC, Murrey DA, Sharp DS, Hitchcock CL, Mojzisik CM, et al. Multimodal imaging and detection strategy with 124 I-labeled chimeric monoclonal antibody cG250 for accurate localization and confirmation of extent of disease during laparoscopic and open surgical resection of clear cell renal cell carcinoma. Surg Innov. 2013;20:59–69.
Mittlmeier LM, Todica A, Gildehaus F-J, Unterrainer M, Beyer L, Brendel M, et al. 68Ga-EMP-100 PET/CT—a novel ligand for visualizing c-MET expression in metastatic renal cell carcinoma—first in-human biodistribution and imaging results. Eur J Nucl Med Mol Imaging. 2022;49:1711–20.
Gnesin S, Müller J, Burger IA, Meisel A, Siano M, Früh M, et al. Radiation dosimetry of 18F-AzaFol: a first in-human use of a folate receptor PET tracer. EJNMMI Res. 2020;10:32.
Kratochwil C, Flechsig P, Lindner T, Abderrahim L, Altmann A, Mier W, et al. 68Ga-FAPI PET/CT: tracer uptake in 28 different kinds of cancer. J Nucl Med. 2019;60:801–5.
Collarino A, Florit A, Bizzarri N, Lanni V, Morganti S, De Summa M, et al. Radioguided surgery with β decay: a feasibility study in cervical cancer. Phys Med. 2023;113:102658.
Alunni Solestizi L, Amoruso R, Biasini M, Bocci V, Campeggi C, Capotosti A, et al. Feasibility study on the use of CMOS sensors as detectors in radioguided surgery with β−- emitters. Appl Radiat Isot. 2020;165. https://doi.org/10.1016/j.apradiso.2020.109347.
Collamati F, Amoruso R, Servoli L, Alunni Solestizi L, Biasini M, Bocci V, et al. Stability and efficiency of a CMOS sensor as detector of low energy β and γ particles. J Instrum. 2020;15:P11003–P11003.
Solestizi LA, Biasini M, Bocci V, Campeggi C, Capotosti A, Collamati F, et al. Use of a CMOS image sensor for beta-emitting radionuclide measurements. J Instrum. 2018;13:7. https://doi.org/10.1088/1748-0221/13/07/P07003.
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement. This research was supported by an NWO-TTW-VICI grant (TTW 16141), an NWO KIC grant (KICH1.ST03.21.030), and a KWF PPS grant (2022-PPS-1/14852). The research leading to these results has received funding from the European Union—NextGenerationEU through the Italian Ministry of University and Research under PNRR—M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" CUP B53C22004000006 and PNRR—M4C2-I1.5 Project ECS 0000024 Rome Technopole,—CUP B83C22002820006. The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the European Institute of Oncology.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Collamati, F., Morganti, S., van Oosterom, M.N. et al. First-in-human validation of a DROP-IN β-probe for robotic radioguided surgery: defining optimal signal-to-background discrimination algorithm. Eur J Nucl Med Mol Imaging 51, 3098–3108 (2024). https://doi.org/10.1007/s00259-024-06653-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-024-06653-6