Abstract
Purpose
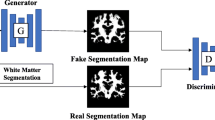
White matter hyperintensities (WMH) are typically segmented using MRI because WMH are hardly visible on 18F-FDG PET/CT. This retrospective study was conducted to segment WMH and estimate their volumes from 18F-FDG PET with a generative adversarial network (WhyperGAN).
Methods
We selected patients whose interval between MRI and FDG PET/CT scans was within 3 months, from January 2017 to December 2018, and classified them into mild, moderate, and severe groups by following the semiquantitative rating method of Fazekas. For each group, 50 patients were selected, and of them, we randomly selected 35 patients for training and 15 for testing. WMH were automatically segmented from FLAIR MRI with manual adjustment. Patches of WMH were extracted from 18F-FDG PET and segmented MRI. WhyperGAN was compared with H-DenseUnet, a deep learning method widely used for segmentation tasks, for segmentation performance based on the dice similarity coefficient (DSC), recall, and average volume differences (AVD). For volume estimation, the predicted WMH volumes from PET were compared with ground truth volumes.
Results
The DSC values were associated with WMH volumes on MRI. For volumes >60 mL, the DSC values were 0.751 for WhyperGAN and 0.564 for H-DenseUnet. For volumes ≤60 mL, the DSC values rapidly decreased as the volume decreased (0.362 for WhyperGAN vs. 0.237 for H-DenseUnet). For recall, WhyperGAN achieved the highest value in the severe group (0.579 for WhyperGAN vs. 0.509 for H-DenseUnet). For AVD, WhyperGAN achieved the lowest score in the severe group (0.494 for WhyperGAN vs. 0.941 for H-DenseUnet). For the WMH volume estimation, WhyperGAN performed better than H-DenseUnet and yielded excellent correlation coefficients (r = 0.998, 0.983, and 0.908 in the severe, moderate, and mild group).
Conclusions
Although limited by visual analysis, the WhyperGAN based can be used to automatically segment and estimate volumes of WMH from 18F-FDG PET/CT. This would increase the usefulness of 18F-FDG PET/CT for the evaluation of WMH in patients with cognitive impairment.






Similar content being viewed by others
References
Fiford CM, Manning EN, Bartlett JW, Cash DM, Malone IB, Ridgway GR, et al. White matter hyperintensities are associated with disproportionate progressive hippocampal atrophy. Hippocampus. 2017;27(3):249–62.
Liu CK, Miller BL, Cummings JL, Mehringer CM, Goldberg MA, Howng SL, et al. A quantitative MRI study of vascular dementia. Neurology. 1992;42(1):138–43.
Erten-Lyons D, Woltjer R, Kaye J, Mattek N, Dodge HH, Green S, et al. Neuropathologic basis of white matter hyperintensity accumulation with advanced age. Neurology. 2013;81(11):977–83.
Lindemer ER, Greve DN, Fischl B, Augustinack JC, Salat DH. Differential regional distribution of juxtacortical white matter signal abnormalities in aging and Alzheimer’s disease. J Alzheimers Dis. 2017;57(1):293–303.
Brickman AM, Zahodne LB, Guzman VA, Narkhede A, Meier IB, Griffith EY, et al. Reconsidering harbingers of dementia: Progression of parietal lobe white matter hyperintensities predicts Alzheimer's disease incidence. Neurobiol Aging. 2015;36(1):27–32.
Tosto G, Zimmerman ME, Hamilton JL, Carmichael OT, Brickman AM. Alzheimer's disease neuroimaging I. The effect of white matter hyperintensities on neurodegeneration in mild cognitive impairment. Alzheimers Dement. 2015;11(12):1510–9.
Scheltens P, Barkhof F, Leys D, Pruvo JP, Nauta JJ, Vermersch P, et al. A semiquantative rating scale for the assessment of signal hyperintensities on magnetic resonance imaging. J Neurol Sci. 1993;114(1):7–12.
Bouter C, Henniges P, Franke TN, Irwin C, Sahlmann CO, Sichler ME, et al. (18)F-FDG-PET detects drastic changes in brain metabolism in the Tg4–42 model of Alzheimer's disease. Front Aging Neurosci. 2018;10:425.
Foster B, Bagci U, Mansoor A, Xu Z, Mollura DJ. A review on segmentation of positron emission tomography images. Comput Biol Med. 2014;50:76–96.
Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58:101552.
Isola P, Zhu J-Y, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. arXiv e-prints; 2016. pp. arXiv:1611.07004.
Son J, Park SJ, Jung K-H. Retinal Vessel Segmentation in fundoscopic images with generative adversarial networks. arXiv e-prints; 2017. pp. arXiv:1706.09318.
Huo Y, Xu Z, Bao S, Bermudez C, Plassard AJ, Liu J, et al. Splenomegaly segmentation using global convolutional kernels and conditional generative adversarial networks. Medical Imaging 2018: Image Processing; 2018. pp. 1057409.
Das JRP, Pankajakshan V. Brain tumor segmentation using discriminator loss. 2019 National Conference on Communications (NCC); 2019. pp. 1–6.
Chen L, Shen C, Zhou Z, Maquilan G, Albuquerque K, Folkert MR, et al. Automatic PET cervical tumor segmentation by combining deep learning and anatomic prior. Phys Med Biol. 2019;64(8):085019.
Leung KH, Marashdeh W, Wray R, Ashrafinia S, Pomper MG, Rahmim A, et al. A physics-guided modular deep-learning based automated framework for tumor segmentation in PET. Phys Med Biol. 2020. https://doi.org/10.1088/1361-6560/ab8535.
Blanc-Durand P, Van Der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: A full 3D U-net convolutional neural network study. PLoS One. 2018;13(4):e0195798.
Oh KT, Lee S, Lee H, Yun M, Yoo SK. Semantic segmentation of white matter in FDG-PET using generative adversarial network. J Digit Imaging. 2020. https://doi.org/10.1007/s10278-020-00321-5.
Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage. 2015;108:214–24.
Nie D, Wang L, Gao Y, Shen D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. Proc IEEE Int Symp Biomed Imaging. 2016;2016:1342–5.
Fazekas F, Chawluk JB, Alavi A, Hurtig HI, Zimmerman RA. MR signal abnormalities at 1.5 T in Alzheimer's dementia and normal aging. AJR Am J Roentgenol. 1987;149(2):351–6.
Nie B, Liu H, Chen K, Jiang X, Shan B. A statistical parametric mapping toolbox used for voxel-wise analysis of FDG-PET images of rat brain. PLoS One. 2014;9(9):e108295.
Tsai JZ, Peng SJ, Chen YW, Wang KW, Li CH, Wang JY, et al. Automated segmentation and quantification of white matter hyperintensities in acute ischemic stroke patients with cerebral infarction. PLoS One. 2014;9(8):e104011.
Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302.
Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging. 2018;37(12):2663–74.
Guerrero R, Qin C, Oktay O, Bowles C, Chen L, Joules R, et al. White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. NeuroImage: Clin. 2018;17:918–34.
Prins ND, Scheltens P. White matter hyperintensities, cognitive impairment and dementia: an update. Nat Rev Neurol. 2015;11(3):157–65.
Xu TGY, Bloch I. From neonatal to adult brain MR image segmentation in a few seconds using 3D-like fully convolutional network and transfer learning. 2017 IEEE International Conference on Image Processing (ICIP); 2017. pp. 4417–21.
Andermatt S, Pezold S, Cattin P. Multi-dimensional gated recurrent units for the segmentation of biomedical 3D-data. Cham: Springer International Publishing; 2016. p. 142–51.
Li H, Jiang G, Zhang J, Wang R, Wang Z, Zheng WS, et al. Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images. Neuroimage. 2018;183:650–65.
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea and the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Ministry of Science and ICT (NRF-2018M3C7A1056898 and NRF-2018M3A9H6081483).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in the studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Rights and permissions
About this article
Cite this article
Oh, K.T., Kim, D., Ye, B.S. et al. Segmentation of white matter hyperintensities on 18F-FDG PET/CT images with a generative adversarial network. Eur J Nucl Med Mol Imaging 48, 3422–3431 (2021). https://doi.org/10.1007/s00259-021-05285-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-021-05285-4