Abstract
Purpose
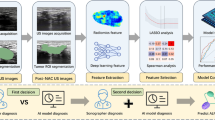
This study proposes an automated prostate cancer (PC) lesion characterization method based on the deep neural network to determine tumor burden on 68Ga-PSMA-11 PET/CT to potentially facilitate the optimization of PSMA-directed radionuclide therapy.
Methods
We collected 68Ga-PSMA-11 PET/CT images from 193 patients with metastatic PC at three medical centers. For proof-of-concept, we focused on the detection of pelvis bone and lymph node lesions. A deep neural network (triple-combining 2.5D U-Net) was developed for the automated characterization of these lesions. The proposed method simultaneously extracts features from axial, coronal, and sagittal planes, which mimics the workflow of physicians and reduces computational and memory requirements.
Results
Among all the labeled lesions, the network achieved 99% precision, 99% recall, and an F1 score of 99% on bone lesion detection and 94%, precision 89% recall, and an F1 score of 92% on lymph node lesion detection. The segmentation accuracy is lower than the detection. The performance of the network was correlated with the amount of training data.
Conclusion
We developed a deep neural network to characterize automatically the PC lesions on 68Ga-PSMA-11 PET/CT. The preliminary test within the pelvic area confirms the potential of deep learning methods. Increasing the amount of training data should further enhance the performance of the proposed method and may ultimately allow whole-body assessments.





Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Maurer T, Eiber M, Schwaiger M, Gschwend JE. Current use of PSMA–PET in prostate cancer management. Nat Rev Urol. 2016;13:226.
Howlader N, Noone A, Krapcho M, Neyman N, Aminou R, Altekruse S, et al. SEER cancer statistics review, 1975-2009 (vintage 2009 populations), National Cancer Institute. Bethesda 2012.
Bernacki KD, Fields KL, Roh MH. The utility of PSMA and PSA immunohistochemistry in the cytologic diagnosis of metastatic prostate carcinoma. Diagn Cytopathol. 2014;42:570–5.
Fizazi K, Faivre L, Lesaunier F, Delva R, Gravis G, Rolland F, et al. Androgen deprivation therapy plus docetaxel and estramustine versus androgen deprivation therapy alone for high-risk localised prostate cancer (GETUG 12): a phase 3 randomised controlled trial. Lancet Oncol. 2015;16:787–94.
Attard G, Parker C, Eeles R, Schroffder F, Tomlins SA, Tannock I, et al. Prostate cancer. Lancet. 2016;387:70–82.
Weineisen M, Schottelius M, Simecek J, Baum RP, Yildiz A, Beykan S, et al. 68Ga-and 177Lu-labeled PSMA I&T: optimization of a PSMA-targeted theranostic concept and first proof-of-concept human studies. J Nucl Med. 2015;56:1169–76.
Afshar-Oromieh A, Holland-Letz T, Giesel FL, Kratochwil C, Mier W, Haufe S, et al. Diagnostic performance of 68 Ga-PSMA-11 (HBED-CC) PET/CT in patients with recurrent prostate cancer: evaluation in 1007 patients. Eur J Nucl Med Mol Imaging. 2017;44:1258–68. https://doi.org/10.1007/s00259-017-3711-7.
Kratochwil C, Bruchertseifer F, Giesel FL, Weis M, Verburg FA, Mottaghy F, et al. 225Ac-PSMA-617 for PSMA-targeted a-radiation therapy of metastatic castration-resistant prostate cancer. J Nucl Med. 2016;57:1941–4.
Kratochwil C, Schmidt K, Afshar-Oromieh A, Bruchertseifer F, Rathke H, Morgenstern A, et al. Targeted alpha therapy of mCRPC: dosimetry estimate of 213Bismuth-PSMA-617. Eur Jo Nucl Med Mol Imaging. 2018;45:31–7.
Rahbar K, Schmidt M, Heinzel A, Eppard E, Bode A, Yordanova A, et al. Response and tolerability of a single dose of 177Lu-PSMA-617 in patients with metastatic castration-resistant prostate cancer: a multicenter retrospective analysis. J Nucl Med. 2016;57:1334–8.
Afshar-Oromieh A, Haberkorn U, Zechmann C, Armor T, Mier W, Spohn F, et al. Repeated PSMA-targeting radioligand therapy of metastatic prostate cancer with 131 I-MIP-1095. Eur J Nucl Med Mol Imaging. 2017;44:950–9.
Hofman MS, Violet J, Hicks RJ, Ferdinandus J, Thang SP, Akhurst T, et al. [177Lu]-PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): a single-centre, single-arm, phase 2 study. Lancet Oncol. 2018;19:825–33.
Eiber M, Fendler WP, Rowe SP, Calais J, Hofman MS, Maurer T, et al. Prostate-specific membrane antigen ligands for imaging and therapy. J Nucl Med. 2017;58:67S–76S.
Bieth M, Krönke M, Tauber R, Dahlbender M, Retz M, Nekolla SG, et al. Exploring new multimodal quantitative imaging indices for the assessment of osseous tumor burden in prostate cancer using 68Ga-PSMA PET/CT. J Nucl Med. 2017;58:1632–7.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436.
Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Zero-echo-time and dixon deep pseudo-CT (ZeDD CT): direct generation of pseudo-CT images for pelvic PET/MRI attenuation correction using deep convolutional neural networks with multiparametric MRI. J Nucl Med. 2018;59:852–8. https://doi.org/10.2967/jnumed.117.198051.
Spuhler KD, Gardus J 3rd, Gao Y, DeLorenzo C, Parsey R, Huang C. Synthesis of patient-specific transmission image for PET attenuation correction for PET/MR imaging of the brain using a convolutional neural network'. J Nucl Med. 2018. https://doi.org/10.2967/jnumed.118.214320.
Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE Deep Learning (DIVIDE) Pseudo-CT Synthesis for Pelvis PET/MR Attenuation correction. J Nucl Med. 2018. https://doi.org/10.2967/jnumed.118.209288.
Ciresan D, Giusti A, Gambardella LM, Schmidhuber J. Deep neural networks segment neuronal membranes in electron microscopy images. Advances in neural information processing systems; 2012. p. 2843-51.
Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015. p. 3431-40.
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention: Springer; 2015. p. 234-41.
Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48.
Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177–95. https://doi.org/10.1016/j.media.2017.12.007.
Corral JE, Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Su1337-deep learning to diagnose intraductal papillary mucinous neoplasms (IPMN) with MRI. Gastroenterology. 2018;154:S-524–S-5.
Bi L, Kim J, Kumar A, Wen L, Feng D, Fulham M. Automatic detection and classification of regions of FDG uptake in whole-body PET-CT lymphoma studies. Comput Med Imaging Graph. 2017;60:3–10. https://doi.org/10.1016/j.compmedimag.2016.11.008.
Xu L, Tetteh G, Lipkova J, Zhao Y, Li H, Christ P, et al. Automated whole-body bone lesion detection for multiple myeloma on 68Ga-pentixafor PET/CT imaging using deep learning methods. Contrast Media Mol Imaging. 2018;2018:11. https://doi.org/10.1155/2018/2391925.
Sironi A, Türetken E, Lepetit V, Fua P. Multiscale centerline detection. IEEE Trans Pattern Anal Mach Intell. 2015;38:1327–41.
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer; 2016. p. 424-32.
Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:150203167. 2015.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9. https://doi.org/10.2967/jnumed.117.202317.
Milletari F, Navab N, Ahmadi S-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 3D Vision (3DV), 2016 Fourth International Conference on: IEEE; 2016. p. 565-71.
Bailey J, Piert M. Performance of 68Ga-PSMA PET/CT for prostate cancer management at initial staging and time of biochemical recurrence. Curr Urol Rep. 2017;18:84.
Hoffmann MA, Miederer M, Wieler HJ, Ruf C, Jakobs FM, Schreckenberger M. Diagnostic performance of 68Gallium-PSMA-11 PET/CT to detect significant prostate cancer and comparison with 18FEC PET/CT. Oncotarget. 2017;8:111073.
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. International MICCAI Brainlesion Workshop: Springer; 2017. p. 287-97.
Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010;4:40–79.
Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
Chollet F. Keras. 2015.
Violet J, Jackson P, Ferdinandus J, Sandhu S, Akhurst T, Iravani A, et al. Dosimetry of (177)Lu-PSMA-617 in metastatic castration-resistant prostate cancer: correlations between pretherapeutic imaging and whole-body tumor dosimetry with treatment outcomes. J Nucl Med. 2019;60:517–23. https://doi.org/10.2967/jnumed.118.219352.
Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, et al. Transfer learning for domain adaptation in mri: Application in brain lesion segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention: Springer; 2017. p. 516-24.
Karki K, Saraiya S, Hugo GD, Mukhopadhyay N, Jan N, Schuster J, et al. Variabilities of magnetic resonance imaging-, computed tomography-, and positron emission tomography-computed tomography-based tumor and lymph node delineations for lung cancer radiation therapy planning. Int J Radiat Oncol Biol Phys. 2017;99:80–9. https://doi.org/10.1016/j.ijrobp.2017.05.002.
Zamboglou C, Carles M, Fechter T, Kiefer S, Reichel K, Fassbender TF, et al. Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate-and high-risk prostate cancer-a comparison study with histology reference. Theranostics. 2019;9:2595.
Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. Aggnet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35:1313–21.
Afshar-Oromieh A, Sattler LP, Steiger K, Holland-Letz T, da Cunha ML, Mier W, et al. Tracer uptake in mediastinal and paraaortal thoracic lymph nodes as a potential pitfall in image interpretation of PSMA ligand PET/CT. Eur J Nucl Med Mol Imaging. 2018;45:1179–87.
Afshar-Oromieh A, Avtzi E, Giesel FL, Holland-Letz T, Linhart HG, Eder M, et al. The diagnostic value of PET/CT imaging with the 68 Ga-labelled PSMA ligand HBED-CC in the diagnosis of recurrent prostate cancer. Eur J Nucl Med Mol Imaging. 2015;42:197–209.
Eiber M, Maurer T, Souvatzoglou M, Beer AJ, Ruffani A, Haller B, et al. Evaluation of hybrid 68Ga-PSMA ligand PET/CT in 248 patients with biochemical recurrence after radical prostatectomy. J Nucl Med. 2015;56:668–74.
Sahlmann C-O, Meller B, Bouter C, Ritter CO, Ströbel P, Lotz J, et al. Biphasic 68 Ga-PSMA-HBED-CC-PET/CT in patients with recurrent and high-risk prostate carcinoma. Eur J Nucl Med Mol Imaging. 2016;43:898–905.
Herlemann A, Wenter V, Kretschmer A, Thierfelder KM, Bartenstein P, Faber C, et al. 68Ga-PSMA positron emission tomography/computed tomography provides accurate staging of lymph node regions prior to lymph node dissection in patients with prostate cancer. Eur Urol. 2016;70:553–7.
Maurer T, Weirich G, Schottelius M, Weineisen M, Frisch B, Okur A, et al. Prostate-specific membrane antigen–radioguided surgery for metastatic lymph nodes in prostate cancer. Eur Urol. 2015;68:530–4.
Pfister D, Porres D, Heidenreich A, Heidegger I, Knuechel R, Steib F, et al. Detection of recurrent prostate cancer lesions before salvage lymphadenectomy is more accurate with 68 Ga-PSMA-HBED-CC than with 18 F-Fluoroethylcholine PET/CT. Eur J Nucl Med Mol Imaging. 2016;43:1410–7.
Hijazi S, Meller B, Leitsmann C, Strauss A, Meller J, Ritter C, et al. Pelvic lymph node dissection for nodal oligometastatic prostate cancer detected by 68Ga-PSMA-positron emission tomography/computerized tomography. Prostate. 2015;75:1934–40.
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. 2014;34:1993–2024.
Dolz J, Gopinath K, Yuan J, Lombaert H, Desrosiers C, Ayed IB. HyperDense-Net: A hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans Med Imaging. 2018;38:1116–26.
Oktay O, Ferrante E, Kamnitsas K, Heinrich M, Bai W, Caballero J, et al. Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE Trans Med Imaging. 2017;37:384–95.
Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Lung and pancreatic tumor characterization in the deep learning era: novel supervised and unsupervised learning approaches. IEEE Trans Med Imaging 2019.
Leclerc S, Smistad E, Pedrosa J, Østvik A, Cervenansky F, Espinosa F, et al. Deep learning for segmentation using an open large-scale dataset in 2d echocardiography. IEEE Trans Med Imaging 2019.
Gibson E, Giganti F, Hu Y, Bonmati E, Bandula S, Gurusamy K, et al. Automatic multi-organ segmentation on abdominal CT with dense v-networks. IEEE Trans Med Imaging. 2018;37:1822–34.
Wang L, Nie D, Li G, Puybareau É, Dolz J, Zhang Q, et al. Benchmark on automatic 6-month-old infant brain segmentation algorithms: the iSeg-2017 challenge. IEEE Trans Med Imaging 2019.
Acknowledgements
We would like to gratefully acknowledge NVIDIA Corporation for the donation of a Titan XP GPU used for this research and thank Professor Paul Cumming for critical reading of the manuscript.
Funding
This study was funded by UniBern Forschungstiftug and Swiss Krebsliga (KFS-4723-02-2019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not describe any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
Supplementary Fig. 1
The lesion-detection performance of the trained model obtained during the cross-validation phase on the test dataset (five-fold cross test). (a) bone lesions, (b) lymph node lesion. (PNG 116 kb)
Supplementary Fig. 2
Test of the influence of the lesion detection threshold on the performance of lesion detection for the proposed triple-combining 2.5D U-Net. (PNG 541 kb)
Rights and permissions
About this article
Cite this article
Zhao, Y., Gafita, A., Vollnberg, B. et al. Deep neural network for automatic characterization of lesions on 68Ga-PSMA-11 PET/CT. Eur J Nucl Med Mol Imaging 47, 603–613 (2020). https://doi.org/10.1007/s00259-019-04606-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04606-y