Abstract
Members of the bacterial phylum Planctomycetota have recently emerged as promising and for the most part untapped sources of novel bioactive compounds. The characterization of more than 100 novel species in the last decade stimulated recent bioprospection studies that start to unveil the chemical repertoire of the phylum. In this study, we performed systematic bioinformatic analyses based on the genomes of all 131 described members of the current phylum focusing on the identification of type III polyketide synthase (PKS) genes. Type III PKSs are versatile enzymes involved in the biosynthesis of a wide array of structurally diverse natural products with potent biological activities. We identified 96 putative type III PKS genes of which 58 are encoded in an operon with genes encoding a putative oxidoreductase and a methyltransferase. Sequence similarities on protein level and the genetic organization of the operon point towards a functional link to the structurally related hierridins recently discovered in picocyanobacteria. The heterologous expression of planctomycetal type III PKS genes from strains belonging to different families in an engineered Corynebacterium glutamicum strain led to the biosynthesis of pentadecyl- and heptadecylresorcinols. Phenotypic assays performed with the heterologous producer strains and a constructed type III PKS gene deletion mutant suggest that the natural function of the identified compounds differs from that confirmed in other bacterial alkylresorcinol producers.
Key points
• Planctomycetal type III polyketide synthases synthesize long-chain alkylresorcinols.
• Phylogenetic analyses suggest an ecological link to picocyanobacterial hierridins.
• Engineered C. glutamicum is suitable for an expression of planctomycete-derived genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the phylum Planctomycetota have gained attention due to their intriguing cellular characteristics. From early research on the phylum starting in 1924 until 2010, only ten species could be brought into axenic culture and have been validly described (Ivanova and Dedysh 2012). Based on intensive worldwide sampling and strain isolation campaigns conducted in the last decade combined with the development of more selective cultivation methods, about 100 novel species have been added to the open strain collection of characterized planctomycetes (Lage and Bondoso 2012; Wiegand et al. 2020, 2018).
Especially Planctomycetia, the class with the currently highest number of described species within the phylum, exhibits biological features different from other members of this phylum (Vitorino and Lage 2022). Dimorphic lifestyles, large genomes of up to 12.4 Mb, high numbers of hypothetical protein-encoding genes, and predicted natural product biosynthetic gene clusters (BGCs) have been described in members of this class (Kallscheuer and Jogler 2021; Kulichevskaya et al. 2017; Overmann et al. 2017). Whereas members of the other classes propagate by binary fission (Wiegand et al. 2020), Planctomycetia members propagate by an uncommon form of asymmetric cell division (often referred to as “budding”).
Due to molecular and genomic insights gained over the last decade, research in the entire phylum is currently in a transition phase from the pure description of novel isolates to an application-based exploration of the biotechnological potential of its members. This is reflected in the recent discovery of the first natural products formed by members of the phylum (Kallscheuer et al. 2020, 2019b; Panter et al. 2019; Sandargo et al. 2020; Santana-Molina et al. 2022). Hitherto, all natural compounds produced by planctomycetes were discovered by untargeted chromatographic analyses. BGCs involved in the production of these empirically confirmed compounds escaped an in silico prediction with bioinformatic tools such as antiSMASH (Blin et al. 2021). In contrast, BGCs belonging to well-characterized compound classes, for example, polyketides and non-ribosomal peptides, can be reliably detected also in planctomycetal genomes. Still, the bioinformatical prediction algorithms are trained with BGCs from well-explored bacterial phyla, for example, actinobacteria or cyanobacteria. This might explain why most of the planctomycetal BGCs do not cluster with BGCs of the MIBiG (Minimum Information about a Biosynthetic Gene cluster) database (Wiegand et al. 2020). This supports the role of planctomycetes as yet for the most part untapped sources of natural products with potential antimicrobial or health-promoting activities (Kallscheuer et al. 2020).
An important class of enzymes involved in natural product biosynthesis are polyketide synthases (PKSs). PKSs use an acyl carrier protein- (ACP) or coenzyme A- (CoA) activated starter unit and catalyze multiple rounds of chain elongation (polyketide formation) using malonyl-CoA as extension unit. Known PKSs are subdivided into three distinct subclasses (types I, II, and III) that reflect different levels of complexity regarding protein domain architectures and catalyzed reaction cascades (Nivina et al. 2019).
In this study, we explore the function of planctomycetal type III PKSs. Enzymes of this class are involved in the biosynthesis of a wide range of bioactive compounds among all domains of life. The best-characterized type III PKSs are plant stilbene and chalcone synthases that catalyze the first committed step of polyphenol biosynthesis pathways (Andrés-Lacueva et al. 2009). Polyphenols, such as stilbenes and flavonoids, show potent health-promoting activities in humans (Pandith et al. 2019; Rozmer and Perjési 2016). Type III PKSs are relatively small enzymes with a length of 250–450 amino acids. They harbor a single active site that typically performs 3–10 rounds of malonyl-CoA-dependent chain elongation and finally cyclize and aromatize the polyketide precursor to a di- or trihydroxylated aromatic ring. In contrast to the other two classes, type III PKSs do not require posttranslational activation by transfer of a phosphopantetheinyl residue to a carrier domain. This facilitates the functional expression of type III PKS-encoding genes in various heterologous hosts. However, the strict regulation of malonyl-CoA biosynthesis and maintenance of only low levels for fatty acid biosynthesis in bacteria turned out to be a major bottleneck for the engineered biosynthesis of polyketide-derived polyphenolic compounds in microorganisms (Lim et al. 2011; Milke et al. 2018; Milke and Marienhagen 2020).
For testing the expression of yet uncharacterized planctomycete-derived type III PKS genes, we employed a previously engineered Corynebacterium glutamicum strain (Milke et al. 2020). Engineered strains of this Gram-positive bacterium are typically used for the production of amino acids (Eggeling and Bott 2015), but in the recent years, various (aromatic) natural compound-producing strains have been constructed (Kogure and Inui 2018; Milke et al. 2020; Wolf et al. 2021).
Materials and methods
Analysis of planctomycetal genomes and construction of phylogenetic trees
The reference genomes of all characterized species of the phylum Planctomycetota were retrieved from GenBank (Table S1). The prediction of BGCs potentially involved in natural product biosynthesis was performed with antiSMASH version 7.0.0 (Blin et al. 2021). The analysis was performed with relaxed strictness and all extra features activated.
For tree construction, the 16S rRNA gene sequences obtained from the planctomycetal genomes were aligned with ClustalW (Thompson et al. 2003). A maximum likelihood phylogenetic tree was calculated from the alignment with FastTree 2.1 (Price et al. 2010) employing the GTR + CAT model and 1000 bootstrap replications. Three 16S rRNA genes of bacterial strains from the Planctomycetota-Verrucomicrobiota-Chlamydiota (PVC) superphylum outside of the phylum Planctomycetota, namely Opitutus terrae (NCBI acc. no. AJ229235), Kiritimatiella glycovorans (acc. no. NR_146840), and Lentisphaera araneosa (acc. no. NR_027571), were used as outgroup.
The locus tags of all planctomycetal type III PKS genes were retrieved from the antiSMASH results, and the encoded protein sequences were extracted from the annotated genomes. Sequences of plant and bacterial type III PKS proteins with known product specificity were obtained from UniProt (or NCBI when the protein was not included in the UniProt database). Alignment and tree construction were performed as described above, but the JTT + CAT model was employed for protein sequences. Phylogenetic trees were visualized with iTOL v6 (Letunic and Bork 2021).
Medium preparation and cultivation
C. glutamicum was routinely cultivated aerobically at 30 °C in brain heart infusion (BHI) medium (Difco Laboratories, Detroit, USA) or defined CGXII medium with 4% (w/v) glucose as sole carbon and energy source (Keilhauer et al. 1993). E. coli DH5α was used for plasmid construction and was cultivated in LB medium (Bertani 1951) at 37 °C. Where appropriate, kanamycin (50 mg/L for E. coli or 15 mg/L for C. glutamicum) and/or spectinomycin (100 mg/L for E. coli and C. glutamicum) was added to the medium. Bacterial growth was followed by measuring the optical density at 600 nm (OD600). C. glutamicum was grown for 6–8 h in test tubes with 5 mL BHI medium on a rotary shaker at 170 rpm (first preculture) and was subsequently inoculated into 50 mL defined CGXII medium with 4% (w/v) glucose in 500 mL baffled Erlenmeyer flasks (second preculture). The cell suspensions were cultivated overnight on a rotary shaker at 130 rpm. The main culture was inoculated to an OD600 of 5.0 in defined CGXII medium with 4% (w/v) glucose. The heterologous episomal gene expression was induced 2 h after inoculation by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG).
Construction of plasmids and strains
All bacterial strains and plasmids used in this study as well as their relevant characteristics are listed in Table 1. Standard protocols of molecular cloning, such as PCR, DNA restriction, and ligation (Sambrook and Russell 2001), were carried out for recombinant DNA work. Techniques specific for C. glutamicum, e.g., electroporation for transformation of strains, were done as described previously (Eggeling and Bott 2005). All enzymes were obtained from ThermoScientific (Schwerte, Germany). Codon-optimized synthetic genes for C. glutamicum ATCC 13032 were obtained from Integrated DNA Technologies (IDT, Leuven, Belgium). Genes were PCR-amplified with the KOD polymerase Master mix (Merck Millipore, Darmstadt, Germany) using primers that introduce flanking homology regions of the acceptor plasmids pMKEx2 or pEKEx3 (Table 2). The PCR products were subsequently used for subcloning into pMKEx2 linearized with BamHI and NcoI using Gibson Assembly. The assembly reaction was incubated for 60 min at 50 °C. In the same manner, PCR products were subcloned into the BamHI/EcoRI-linearized plasmid pEKEx3. All constructed plasmids were finally verified by check-PCR using primers that bind outside of the inserted sequences and by DNA sequencing at Eurofins Genomics (Ebersberg, Germany). The check-PCR was performed with the DreamTaq Green Mastermix (ThermoFisher) with the following program: initial denaturation 5 min, 95 °C; 33 cycles of denaturation (95 °C, 30 s), annealing (57 °C, 20 s), and elongation (72 °C, 1 min/kb), and a final elongation step (95 °C, 5 min).
Construction of gene deletion mutants in Stieleria maiorica.
For the targeted gene inactivation in S. maiorica Mal15T (= DSM 100215 T = LMG 29790 T), the flanking regions up- and downstream of the coding sequence of a specific gene (approximately 1500 bp each) were amplified with the Q5 polymerase Master mix (New England Biolabs) using the recommended standard PCR protocol and subsequently cloned into the plasmid pCJ003x(cat). The amplified homology regions still included the first and last 24 nucleotides of the coding sequence of the inactivation target. In the final plasmid, the inserted homology arms flank the chloramphenicol resistance gene cat including its promoter. Prior to the transformation of S. maiorica, 2 µg of plasmid DNA was linearized with a single-cutting restriction enzyme that cleaves outside of the assembled insert region (upstream homology region–Pcat-cat–downstream homology region) and purified with the NucleoSpin Gel and PCR Clean‑up Kit (Macherey–Nagel, Düren, Germany).
Electrocompetent cells of S. maiorica Mal15T were prepared from 200 mL of an exponentially growing culture (OD600 of 0.5–0.7) in M3H NAG ASW medium. The medium composition was published in a previous study (Sandargo et al. 2020). The cells were harvested by centrifugation (4500 rpm, 4 °C, 20 min) and washed twice with sterile 10% (v/v) glycerol. Cells obtained after the second washing step were mixed with the linearized plasmid DNA and transferred to a Gene Pulser Electroporation Cuvette with 0.2 cm gap (Biorad). The electroporation was performed with a GenePulser Xcell (Biorad) with the following parameters: 2500 V, 25 µF, 200 Ω, pulsing time 5 ms. The cell suspension was immediately transferred to 4 mL M3H NAG ASW medium and cultivated for 2 h at 28 °C on a rotary shaker. The cells were harvested by centrifugation (4500 rpm, 20 °C, 6 min) and ultimately streaked on M3H NAG ASW plates containing 34 mg/L chloramphenicol that were incubated at 28 °C. Colonies obtained after 7–10 days were re-streaked and checked for the expected gene inactivation by PCR (DreamTaq Green Master mix).
Two-phase liquid–liquid extraction of the culture broth and GC-TOF–MS analysis
Extracts of the fermented culture broth (medium and cells) were prepared using a two-phase extraction protocol (Kallscheuer et al. 2016). Briefly, per strain six times 1 mL of culture broth was mixed with the same volume of ethyl acetate. The mixture was shaken in an Eppendorf Thermomixer (1200 rpm, 10 min., 20 °C). After a centrifugation step (13,000 rpm, 6 min, room temperature), 800 µL of the upper ethyl acetate phase was transferred to a solvent-resistant deep-well plate and the solvent was evaporated to dryness in a fume hood overnight. On the next day, the dried extract was pooled in 150 µL acetonitrile per culture and stored at − 20 °C until further processing.
For GC-TOF–MS analysis (Paczia et al. 2012), 130 µL aliquots of the samples were lyophilized overnight in a Christ LT-105 freeze drier (Martin Christ Gefriertrocknungsanlagen). The dried samples were consecutively derivatized with 50 µL 20 mg/mL O-methylhydroxylamine (MeOX) (Sigma Aldrich) in pyridine (Honeywell) for 90 min at 30 °C and 600 rpm in an Eppendorf Thermomixer followed by an incubation with additional 80 µL of MSTFA (N-acetyl-N-(trimethylsilyl)-trifuoroacetamide) (Sigma-Aldrich) for 90 min at 40 °C and 600 rpm. For the determination of the derivatized metabolites, an Agilent 8890N double SSL gas chromatograph (Agilent, Waldbronn, Germany) equipped with an L-PAL3-S15 liquid autosampler was used, coupled to a LECO Pegasus GC-HRT + 4D high-resolution time of flight mass spectrometer (LECO, Mönchengladbach, Germany). The system was controlled by the LECO ChromaToF software. For 1D GC analysis, the back injector was equipped with a 30 m Agilent EZ-Guard VF-5 ms + 10 m guard column (Agilent, Waldbronn, Germany). The front injector was equipped with a 30 m HP 5-ms Ui column (HP) connected to a 2 m Rtxi17 (Restek) in a secondary oven. A constant helium flow was adjusted to 1 mL/min for the active (back) injector and column and to 0.5 mL/min for the passive (front) injector. The GC temperature program started at 60 °C with a hold time of 2 min, followed by a temperature ramp of + 12 °C/min up to the final temperature of 300 °C, hold time of 8 min. This leads to a total run time of 30 min. The secondary oven temperature offset was set to 15 °C above the first oven temperature. The transfer line temperature was set to 300 °C. The TOF–MS was operated in positive electron impact [EI]+ mode at an electron energy of 70 eV. The ion source temperature was set to 250 °C. The mass spectrometer was tuned and calibrated with the mass fragmentation pattern of PFTBA (perfluorotributylamine). During analysis, the accurate masses were corrected to a single point lockmass of Heptacosa as an external reference at m/z 218.9856. 1D Data acquisition was performed in stick mode with a rate of 12 scans/s. To annotate known metabolites, a baseline noise-corrected fragmentation pattern together with the corresponding current RI value (Retention time Index) was compared to the in-house accurate m/z database JuPoD and the commercial nominal m/z database NIST20 (National Institute of Standards and Technology, USA). Unknown peaks were identified by a virtual reconstruction of the derivatized metabolite structure via the measured baseline noise-corrected accurate mass m/z fragment pattern in comparison to an accurate m/z fragment register inside the JuPoD main library and were subsequently verified by virtual derivatization and fragmentation of the predicted structure.
Antimicrobial activity assay
For testing a potential antimicrobial activity of the compounds produced by planctomycetal type III PKSs in C. glutamicum, the constructed strains were cultivated as described above. Three days after induction of the episomal gene expression, the entire culture volume (50 mL per strain) was extracted using the two-phase extraction protocol with ethyl acetate described above. The up-scaled procedure was performed in 50 mL Falcon tubes. After the centrifugation step, the organic phase was collected in a round bottom flask and the ethyl acetate was evaporated in a rotary evaporator (Rotavapor R-114 connected to a B-480 water bath, Büchi) at 40 °C and 240 mbar. Each extract was evaporated to dryness and resuspended in 200 µL methanol. Subsequently, 20 µL of the resuspended extract was added to a filter disk (6 mm) and allowed to dry for ca. 20 min. The disks were placed on LB plates inoculated with the indicator strains E. coli ∆tolC (Donner et al. 2017) and Bacillus subtilis DSM10T. The plates were incubated at 28 °C. Discs with kanamycin (10 µg per disc) were used as a positive control, while 20 µL methanol and 20 µL sterile ddH2O served as negative controls. The plates were inspected for zones of inhibition after 24 h of incubation.
β-Lactam antibiotic sensitivity assays
To check for an increased resistance of the constructed type III PKS gene-expressing C. glutamicum strains to β-lactam antibiotics, the empty vector control strain and the eight expression strains were cultivated as described above. Three days after the induction of the episomal expression of planctomycete-derived genes, 100 µL of the culture was streaked on CGXII plates supplemented with 4% (w/v) glucose, 1 mM IPTG, 15 mg/L kanamycin (and 100 mg/L spectinomycin for the strains harboring two plasmids). Two ampicillin minimal inhibitory concentration (MIC) test stripes (0.016–256 μg/mL, Bestbion dx) were placed on each plate, and all plates were incubated for 2 days at 28 °C. The MIC values were visually determined from the obtained zones of inhibition. The constructed gene inactivation mutants of S. maiorica Mal15T were tested for sensitivity to ampicillin using the same protocol but were streaked on M3H NAG ASW plates with 34 mg/L chloramphenicol and incubated at 28 °C for 2 days.
Results
Distribution of type III polyketide synthase genes in the phylum Planctomycetota.
The type strain genomes of all currently described species of the phylum Planctomycetota were analyzed for the presence of type III PKS-encoding genes using the antiSMASH pipeline. The current collection of effectively or validly described members of the phylum includes 137 species (as of June 2023). The genomes of the type strains of six species have not been sequenced yet. The analysis of 131 genomes (Table S1) uncovered that 91 genomes encode at least one type III PKS. A single strain, Thalassoroseus polymorphus Mal48T, carries two type III PKS genes (Rivas-Marin et al. 2020), whereas even three type III PKS genes were found in two strains, Lacipirellula parvula PX69T and Lacipirellula limnantheis I41T (Dedysh et al. 2020; Kallscheuer et al. 2021). With the presence of type III PKS genes in 70% of the known species (91 out of 131 planctomycetal genomes), these genes are found in a majority of the phylum, but are not conserved in all species.
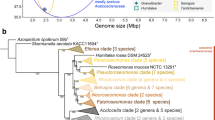
As visualized in the phylogenetic tree, the presence or absence of type III PKS genes cannot be clearly attributed to the individual bacterial families (Fig. 1). However, their relative abundance is lower in the classes Phycisphaerae and Candidatus Brocadiae. This finding aligns with the observation that members of these classes have smaller genomes and harbor lower numbers of BGCs than members of the class Planctomycetia (Kallscheuer and Jogler 2021).
16S rRNA gene phylogenetic tree. A maximum likelihood phylogenetic tree was calculated based on the 16S rRNA gene sequences of the type strains of the currently described species within the phylum Planctomycetota. Sequences of three members of the PVC superphylum were used as outgroup (Opitutus terrae, NCBI acc. no. AJ229235; Kiritimatiella glycovorans, acc. no. NR_146840; and Lentisphaera araneosa, acc. no. NR_027571). The scale bar indicates the number of substitutions per nucleotide position. The presence of at least one type III polyketide synthase-encoding gene in the genome of a strain is indicated by a light green box next to the species name, whereas the absence of such genes is indicated by a red box. Gray boxes indicate species for which the genome has not been sequenced yet. A dark green box indicates the organization of the type III PKS gene in a three-gene operon with a methyltransferase and an oxidoreductase gene
The encoded putative type III PKS proteins cover a size range from 274 aa (Botrimarina mediterranea, locus tag Spa11_43050) to 441 aa (Rubripirellula amarantea, locus tag Pla22_34060) and have an average size of 371 aa. All candidate sequences are either annotated as alpha-pyrone synthesis polyketide synthase, type III polyketide synthase, or chalcone and stilbene synthase domain-containing proteins. The compounds synthesized by these proteins are unknown. All planctomycetal type III PKSs harbor the N-terminal and C-terminal domains found in plant chalcone and stilbene synthases (Pfam domains PF00195 and PF02797).
Of the analyzed set of 96 sequences, two did not cluster with all other sequences. These belong to the type strains of the two species that turned out to have three type III PKS genes (L. parvula PX69T and L. limnantheis I41T) (Fig. 2, next to the outgroup). In a clustering analysis based on all-against-all BLAST + similarities, these two sequences did not cluster with any characterized type III PKS of bacterial, fungal, or plant origin (data not shown).
Phylogenetic tree based on type III polyketide synthase protein sequences. The maximum likelihood phylogenetic tree was calculated based on the protein sequences of all putative type III PKSs in members of the phylum Planctomycetota. The sequences of citrate synthase of C. glutamicum (Uniprot-ID P42457) and FtsZ of E. coli K12 (Uniprot-ID P0A9A6) were used as outgroup. Bootstrap values obtained after 1000 re-samplings were calculated, and values of > 90% are indicated by open circles on the branches. The circle size reflects the bootstrap values between 90% (smaller circles) and 100% (larger circles). Colored boxes next to the species name indicate members belonging to the same family or class. The larger open circles next to the species names indicate that the protein was selected for a heterologous expression of the respective gene in C. glutamicum
To gain more insights into the potential compound class produced by planctomycetal type III PKSs, we also analyzed the organization of neighboring genes encoded immediately up- and downstream. In 60% of the cases (58 of the 96 analyzed genes), the type III PKS gene turned out to be the last gene of a putative tricistronic operon, in which the PKS gene is preceded by a putative methyltransferase- and FAD-dependent oxidoreductase gene (Fig. 1, dark green boxes).
Genetic organization of type III PKS genes and selection of candidates for heterologous expression experiments
Together with the phylogenetic distance of the donor strains, we considered the information on the genetic organization during selection of genes for the planned heterologous expression in C. glutamicum. Six putative type III PKSs from species belonging to different families were selected for a heterologous expression of the respective genes codon-optimized for C. glutamicum. The donor planctomycetes included the limnic model strain Planctopirus limnophila DSM 3776 T, the marine model strain Rhodopirellula baltica SH1T, the first isolated planctomycete strain Gimesia maris DSM 8797 T, and strains isolated within the recent years, Stieleria maiorica Mal15T, Alienimonas californiensis CA12T, and “Saltatorellus ferox” Poly30T (a more distantly related strain probably belonging to a separate phylum). S. maiorica was included because of its genetical accessibility.
An analysis of the genomic context showed that G. maris and R. baltica harbor the three gene operons (methyltransferase – oxidoreductase – type III PKS), whereas S. maiorica harbors a two-gene operon lacking the putative oxidoreductase gene (Fig. 3). In “S. ferox,” a putative pentacistronic operon was found. In this organism, the type III PKS gene is preceded by two putative methyltransferase-encoding genes that are in turn preceded by genes encoding a putative lipoprotein and lipid/isoprenoid-binding protein. The genes from A. californiensis and P. limnophila are probably not part of an operon, and neither methyltransferase nor oxidoreductase genes are encoded in direct proximity (Fig. 3).
Genetic organization of the type III PKS genes selected for heterologous expression experiments in C. glutamicum. The type III PKS-encoding gene and the adjacent genes in six selected planctomycetal strains are depicted. Same colors indicate the same putative gene function: methyltransferase (turquoise), oxidoreductase (orange), type III polyketide synthase (grey-blue and blue, see text for details), putative lipoproteins (green). For the other genes (grey), the annotation is not specified here
Heterologous expression of planctomycetal genes in C. glutamicum.
The strain C. glutamicum M-CoA was used as a host for heterologous expression experiments (Milke et al. 2019, 2020). This strain was extensively engineered towards aromatic compound (in particular polyphenol) biosynthesis by the implementation of several genomic modifications (Milke et al. 2020), including the abolishment of all known catabolic pathways for aromatic compounds naturally present in the wild-type strain (Kallscheuer and Marienhagen 2018; Kallscheuer et al. 2016). In addition, the central metabolism of the strain was engineered to redirect the carbon flux towards malonyl-CoA, the rate-limiting metabolite for engineered biosynthesis of polyketide-derived phenolic compounds (Lim et al. 2011; Milke et al. 2019). The strain turned out to be suitable for the biosynthesis of polyketide-derived compounds from plants and fungi (Kallscheuer et al. 2019a, 2017). It also harbors the T7 RNA polymerase-encoding gene under the control of an IPTG-inducible promoter that allows the inducible expression of genes placed under the control of the strong T7 promoter (Kortmann et al. 2015). Genes coding for the six selected type III PKSs from planctomycetes (Fig. 3) were purchased as codon-optimized synthetic DNA fragments and cloned into the plasmid pMKEx2 allowing an expression under control of the strong T7 promoter.
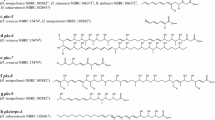
The constructed C. glutamicum strains, each expressing a single type III PKS gene, were cultivated in defined CGXII medium with 4% (w/v) glucose as the sole carbon and energy source under inducing conditions. An uninduced plasmid-free strain and a strain harboring the empty vectors (pMKEx2 and pEKEx3) were used as negative controls (Fig. S1A, B). All seven strains reached the same final OD600 of 42–45 after 72 h. The cultures were then harvested, and extracts were subjected to an untargeted GC-TOF–MS analysis. No products were detected for the episomal expression of the codon-optimized genes coding for the enzymes from P. limnophila (t3pksPlCg) and A. californiensis (t3pksAcCg) (that both do not harbor the three gene operons or variations thereof) (Fig. S1C, E). However, for the strains with enzymes from G. maris (t3pksGmCg), R. baltica (t3pksRbCg), and S. maiorica (t3pksSmCg), additional peaks not present in the empty vector control were detected (Fig. S1D, F, H). A comparison of the mass spectra against the NIST database and the in-house database JuPoD identified the compounds as alkylresorcinols (Table 3). Their identity was confirmed by virtual derivatization and electron impact (EI+) fragmentation of the predicted structure (Figs. S3-S8). Alkylresorcinols are derivatives of 1,3-dihydroxybenzene (resorcinol) that harbor an additional alkyl chain at C5 of the aromatic ring (in meta position to both hydroxy groups) (Fig. 4). This ring substitution pattern resembles compounds produced by other type III polyketide synthases, e.g., stilbene synthases (Parage et al. 2012).
Postulated alkylresorcinol pathway in members of the phylum Planctomycetota and Hierridin biosynthesis pathway in Cyanobium sp. LEGE 06113. The reaction steps leading to alkylresorcinols including relevant co-substrates and products are depicted for the postulated alkylresorcinol pathway in members of the phylum Planctomycetota. The Hierridin biosynthetic pathway identified in Cyanobium sp. LEGE 06113 is shown for comparison. n = 14 or 16 for the planctomycetal pathway and n = 14 for the picocyanobacterial pathway. SAM, S-adenosyl methionine; SAH, S-adenosyl homocysteine
Based on the C2-C7 aldol condensation mechanism performed by stilbene synthases (Bhan et al. 2015), a putative biosynthesis pathway for the identified pentadecyl- and heptadecylresorcinols was outlined. In this pathway, the three alkylresorcinol-producing enzymes from G. maris, R. baltica, and S. maiorica use CoA-activated long-chain fatty acids as starter units and catalyze an iterative and decarboxylating condensation of three molecules of malonyl-CoA for production of a tetraketide intermediate that is subsequently cyclized and aromatized to a resorcinol residue (Fig. 4A, upper branch). CoA-activated stearic acid (18:0) and oleic acid (18:1cis-9) are the required starter units to produce 5-heptadecylresorcinol and 5-heptadecenylresorcinol, respectively (Table 3). The enzymes from G. maris and S. maiorica probably also accepted palmitoyl-CoA (16:0) as substrate to produce 5-pentadecylresorcinol. The three free fatty acids (16:0, 18:0, and 18:1cis-9) were also identified in the GC-TOF–MS analysis of the constructed C. glutamicum strains (Fig. S1). Hence, the suspected biosynthetic route appears plausible.
The enzyme from “S. ferox” also produced the three above-mentioned compounds (Fig. S1G); however, peaks with a similar fragmentation pattern in the EI+ spectrum but a mass increase of 14 Da were detectable. Moreover, in silico-fragmentation suggests an additional methyl group attached to the aromatic ring (Figs. S6-8). The most likely explanation for this observation is that the enzyme from “S. ferox” can incorporate the alternative extender unit methylmalonyl-CoA in one of the three chain elongation steps (Fig. 4A, lower branch).
The obtained results suggest that planctomycetal type III PKSs organized in the three gene operons (or variations thereof) act as alkylresorcinol synthases. To investigate the role of the other, in the natural host co-occurring genes, we constructed plasmids based on the vector pEKEx3 that is compatible with pMKEx2 in C. glutamicum. Codon-optimized genes encoding the putative FAD-dependent oxidoreductase (fdo) and methyltransferase (mtr) from G. maris were inserted as bicistronic operon into the multiple cloning site of pEKEx3 to yield pEKEx3_mtrGmCg-fdoGmCg and the two putative methyltransferase genes from “S. ferox” were used for the construction of pEKEx3_mtr1SfCg-mtr2SfCg. Cultures of the strains C. glutamicum M-CoA pMKEx2_t3pksGmCg pEKEx3_mtrGmCg-fdoGmCg and C. glutamicum M-CoA pMKEx2_t3pksSfCg pEKEx3_mtr1SfCg-mtr2SfCg were prepared and processed as described for the previous experiment. The co-expression of the two additional genes derived from G. maris led to a single methylation in the alkylresorcinols (Fig. S2). The two hydroxy groups of the alkylresorcinols are shielded with trimethylsilyl (TMS) residues after compound derivatization prior to GC-TOF–MS analysis. Since only alkylresorcinol compounds with two TMS residues were identified, the additional methyl group is probably immediately connected to a carbon atom and not to an oxygen atom. However, the exact position of the C-methylation on the aromatic ring could not be determined from the fragmentation pattern. Taken together, the role of the putative FAD-dependent oxidoreductase from G. maris and the two methyltransferases from “S. ferox” Poly30T remain to be elucidated. The identified alkylresorcinols produced by the planctomycetal type III PKSs are likely the first intermediate of an at least three-step pathway.
Comparative analysis with bacterial and plant type III PKSs participating in explored biosynthesis pathways
To check for a potential protein sequence-function relationship, a maximum likelihood phylogenetic tree was constructed based on the six investigated planctomycetal enzymes and other plant or bacterial type III PKSs participating in already explored biosynthesis pathways. The type III PKSs used for comparison included model stilbene synthases (STS), chalcone synthases (CHS) and pentaketide chromone synthases (PCS) from plants, and synthases for 1,3,6,8-tetrahydroxynaphthalene (TNHS, RppA), phloroglucinol (PhlD), 3,5-dihydroxyphenylacetyl-CoA (DpgA), resorcinols (AgqA, SrsA), and α-pyrones (PKS11) from bacteria (Liu and Begley 2020). The clustering of type III PKSs (Fig. 5) reflects a possible sequence-function relationship as distinct clades were obtained for synthases that form the same compound classes. Similarly, methylmalonyl-CoA-incorporating PKSs did not cluster with their malonyl-CoA-specific counterparts. Five of the planctomycetal enzymes clustered within the clade of malonyl-CoA-specific enzymes, whereas the enzyme from “S. ferox” clustered within the clade of methylmalonyl-CoA-incorporating PKSs (Fig. 5). This clustering pattern is in line with the finding that (from the tested enzymes) only the type III PKS from “S. ferox” formed alkylresorcinols bearing an additional methyl group that potentially results from the incorporation of methylmalonyl-CoA in the growing polyketide chain. The other five tested planctomycetal synthases cluster on a separate branch with the type III PKS HidC from the picocyanobacterium Cyanobium sp. LEGE 06113 (Costa et al. 2019). The position of HidC in the phylogenetic tree was confirmed by the calculation of a protein sequence similarity matrix based on the alignment that was also used for tree construction (Table S2). Indeed, HidC showed higher protein sequence identity values to the enzymes from G. maris (54.8%) and S. maiorica (50.6%) than these two to the other tested planctomycetal type III PKSs (< 48%). The type III PKS HidC is not only an alkylresorcinol synthase that formed the same compounds as the closely related proteins tested from planctomycetes, but also the last gene in a three-gene operon preceded by a methyltransferase (HidA) and an oxidoreductase (HidB) (Costa et al. 2019).
Phylogenetic tree of the investigated planctomycetal type III PKS and enzymes of this class with an identified reaction product from other organisms. The tree was constructed based on an alignment of type III PKS protein sequences. The sequences of citrate synthase of C. glutamicum (Uniprot-ID P42457) and FtsZ of E. coli K12 (Uniprot-ID P0A9A6) were used as outgroup. Bootstrap values obtained after 1000 re-sampling are shown on the nodes (in %). The Uniprot-ID of the used proteins are provided in brackets. In case that a protein was not listed in the Uniprot database, the NCBI accession numbers is presented alternatively (in italics)
Analysis of potential functions of bacterial alkylresorcinols
Alkylresorcinols and decorated phenolic lipids derived thereof are amphiphilic compounds that have been identified across the bacterial kingdom (Zabolotneva et al. 2022). However, their natural functions are only occasionally understood. Cyanobacteria produce several alkylresorcinols with antibiotic activities (Martins et al. 2019). The actinomycete Actinoplanes missouriensis uses a type III PKS (AgqA) to produce alkylresorcinols, which are subsequently prenylated and hydroxylated (Awakawa et al. 2011); however, their function remains enigmatic. Streptomyces griseus, a member of the same phylum, and Myxococcus xanthus (class Deltaproteobacteria) further oxidize alkylresorcinols produced by type III PKSs (SrsA and FtpA, respectively) to alkylquinones (Funabashi et al. 2008; Hayashi et al. 2011). The inactivation of srsA in S. griseus led to an increased sensitivity to β-lactam antibiotics indicating that alkylquinones confer an increased resistance to this class of antibiotics (Funabashi et al. 2008). In Mycobacterium tuberculosis, alkylquinones (polyketide quinones) were shown to act as intermediate electron carriers during respiration in oxygen-deficient niches (Anand et al. 2015).
To deduce potential functions of alkylresorcinols in planctomycetes, the genetically accessible S. maiorica Mal15T and the constructed C. glutamicum strains were subjected to further studies. An inactivation of the sole type III PKS-encoding gene in S. maiorica (locus tag Mal15_23020) was achieved by enforcing a simultaneous homologous recombination event up- and downstream of the open reading frame of Mal15_23020. The gene was exchanged for a heterologous DNA fragment that encodes a chloramphenicol acetyltransferase (cat) gene including its promoter. As a control, we also inactivated the gene Mal15_51000 that encodes a non-essential protein for which a deletion did not lead to a detectable phenotype under standard cultivation conditions. The recombinant strains S. maiorica ∆Mal15_23020::cat and S. maiorica ∆Mal15_51000::cat were cultivated in M3H NAG ASW medium with chloramphenicol; however, no differences in growth speed or final biomass yield were observed. Hence, alkylresorcinol biosynthesis is not essential in S. maiorica under the tested growth conditions.
To test for an influence of alkylresorcinol biosynthesis on the natural resistance of S. maiorica to β-lactam antibiotics, both strains were streaked on M3H NAG ASW agar with chloramphenicol and minimal inhibitory activity (MIC) test stripes with ampicillin (0.016–256 μg/mL) were placed on the plates. Both strains grew equally well up to the highest tested ampicillin concentration (Fig. S9). Hence, alkylresorcinols do not play a role as a resistance mechanism to β-lactam antibiotics in planctomycetes. This observation is in line with the presence of β-lactamases as resistance-conferring factors in planctomycetes and is additionally substantiated by the fact that also planctomycetal strains that lack type III PKS-encoding genes are resistant to β-lactam antibiotics. Similarly, cell material from the main cultures of the C. glutamicum strains producing planctomycetal alkylresorcinols was streaked on CGXII agar with 4% glucose (w/v), 1 mM IPTG and the respective antibiotics for plasmid maintenance (kanamycin and/or spectinomycin). Ampicillin MIC test stripes yielded similar MICs between 1.5 and 2 µg/mL for all tested strains and provided no indications for an increased resistance (Fig. S10). Since the heterologous planctomycetal enzymes did not oxidize the produced compounds to alkylquinones in C. glutamicum, a similar effect as observed for alkylquinones in S. griseus cannot be excluded at this stage. It should also be noted that the alkylresorcinols found in the constructed C. glutamicum strains are not necessarily the identical (final) compounds produced in the planctomycetes.
Culture extracts of the constructed C. glutamicum strains were used for agar disc diffusion assays to test for antimicrobial activities. Unexpectedly, zones of inhibition were obtained for all cultures including the empty vector control (Fig. S11). However, these resulted from the toxicity of the inducer IPTG in the concentrated extract. An additional antimicrobial effect of the alkylresorcinol core structures formed in C. glutamicum was not observed. The exact function of the compounds remains elusive.
Discussion
Bioinformatic genome mining of the current open collection of 131 planctomycetal species with a sequenced genome yielded 96 putative type III PKS genes. The phylogenetic tree based on the encoded protein sequences did not strictly reflect the phylogenetic relationship of the strains to which they belong (Fig. 2). However, proteins from species belonging to the same genus clustered in most cases. It has been reported that the phylogeny of biosynthetic enzymes shows a cluster pattern according to their catalytic function and rapidly deviates from the phylogeny of their host strains (Medema et al. 2014), explaining the observed low resolution of the type III PKS protein sequences when used as phylogenetic marker.
Fifty-eight of the analyzed strains harbor a conserved three-gene operon consisting of a putative methyltransferase-, an oxidoreductase-, and a type III PKS-encoding gene. The cluster is spread across different families within the phylum and strains from various habitats (freshwater, seawater, and soil). An exemplary empirical analysis of four of these clusters (two of them with variations of the three gene operons) confirms their relevance for the biosynthesis of alkylresorcinols. The type III PKSs of all tested strains except “S. ferox” clustered with HidC of Cyanobium sp. LEGE 06113. The gene hidC is part of the more recently identified three-gene operon hidABC that is responsible for the production of hierridin B, a 5-pentadecylresorcinol derivative that is additionally hydroxylated at position 4 and O-methylated at positions 1 and 3 of the phenolic residue (Costa et al. 2019) (Fig. 4B). Hierridins were already identified in the early 1990s in the lichen Ramalina farinacea and in the cyanobacterium Phormidium ectocarpi (Gonzalez et al. 1992; Papendorf et al. 1998). Ten years ago, additional hierridins were discovered in filamentous and unicellular marine cyanobacteria and anti-tumor activities were reported (Leão et al. 2013). Hierridin B was recently used as a basis to synthesize chemical analogues that turned out to have potent anti-proliferative activity (Brandão et al. 2020).
The hidABC cluster is found in many cyanobacterial genomes, and a close relationship to the here investigated cluster in members of the phylum Planctomycetota was already observed by the authors of the hierridin study. The order of the genes in the operon (methyltransferase–oxidoreductase–type III PKS) is conserved in nearly all analyzed planctomycetes in which the operon is present and identical to the cyanobacterial hidABC operon. On protein level, all three encoded proteins show a similar degree of sequence identity between 48 and 55% to the cyanobacterial counterparts. While the planctomycetal type III polyketide synthases synthesized the same major product as HidC, the specificity of the methyltransferases differs. The enzyme of G. maris is probably a C-methyltransferase, whereas HidA methylates the two phenolic hydroxy groups of the alkylresorcinol (Costa et al. 2019). The high degree of relationship in terms of genetic organization, enzymatic activity, and protein sequence identity raises the question if the cluster was transferred from one phylum to the other via horizontal gene transfer (Meslet-Cladière et al. 2013). The phyla Cyanobacteriota and Planctomycetota are only distantly related (Hug et al. 2016); hence, it appears rather unlikely that the cluster was already present in a common ancestor. The horizontal gene transfer hypothesis receives further support when considering that planctomycetes are often associated with aquatic phototrophs (Kallscheuer et al. 2021; Lage and Bondoso 2014). Genes up- and downstream of the planctomycetal operon are not conserved compared to Cyanobium sp. LEGE 06113. Adjacent genes are also not conserved among the analyzed planctomycetal genomes, but only in close relatives of a respective strain. The operon is widely distributed in cyanobacteria, and the analyzed Cyanobium sp. (or an ancestor) is hence not necessarily the donor/acceptor of the transfer event. The hypothesis further substantiates suspected planctomycete-cyanobacterial allelopathic interactions and thus deserves further attention.
Since the recombinant C. glutamicum strains only produced the alkylresorcinol core, but not the fully decorated compounds suspected in planctomycetes, an elucidation of the natural function is hampered by current limitations of the used heterologous expression approach. Still, the acquired knowledge on the chemical nature of the identified compounds allows for a targeted search in planctomycetal extracts in future studies. The results also indicate that alkylresorcinols are not the only compounds produced by planctomycetal type III PKS. Structurally more unrelated enzymes in strains harboring more than one type III PKS and genes not part of the characterized operon are probably only a small fraction of the yet for the most part untapped chemical repertoire of the phylum Planctomycetota. Bioprospection in the phylum will be guided by the continuous exploration of the phylogenetic diversity and the development of more sophisticated genetic tools.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Anand A, Verma P, Singh Anil K, Kaushik S, Pandey R, Shi C, Kaur H, Chawla M, Elechalawar Chandra K, Kumar D, Yang Y, Bhavesh Neel S, Banerjee R, Dash D, Singh A, Natarajan Vivek T, Ojha Anil K, Aldrich Courtney C, Gokhale Rajesh S (2015) Polyketide quinones are alternate intermediate electron carriers during mycobacterial respiration in oxygen-deficient niches. Mol Cell 60(4):637–650. https://doi.org/10.1016/j.molcel.2015.10.016
Andrés-Lacueva et al., 2009 Andrés‐Lacueva C, Medina‐Remon A, Llorach R, Urpi‐Sarda M, Khan N, Chiva‐Blanch G, Zamora‐Ros R, Rotches‐Ribalta M, Lamuela‐Raventós RM (2009) Phenolic compounds: chemistry and occurrence in fruits and vegetables. In Fruit and vegetable phytochemicals (eds L.A. de la Rosa, E. Alvarez-Parrilla and G.A. González-Aguilar). doi:10.1002/ 9780813809397.ch2
Awakawa T, Fujita N, Hayakawa M, Ohnishi Y, Horinouchi S (2011) Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. ChemBioChem 12(3):439–448. https://doi.org/10.1002/cbic.201000628
Bertani G (1951) Studies on lysogenesis I: the mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62(3):293
Bhan N, Li L, Cai C, Xu P, Linhardt RJ, Koffas MAG (2015) Enzymatic formation of a resorcylic acid by creating a structure-guided single-point mutation in stilbene synthase. Protein Sci 24(2):167–173. https://doi.org/10.1002/pro.2600
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema Marnix H, Weber T (2021) antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49(W1):W29–W35. https://doi.org/10.1093/nar/gkab335
Brandão P, Moreira J, Almeida J, Nazareth N, Sampaio-Dias IE, Vasconcelos V, Martins R, Leão P, Pinto M, Saraíva L, Cidade H (2020) Norhierridin B, a new hierridin B-based hydroquinone with improved antiproliferative activity. Molecules 25(7):1578
Costa M, Sampaio-Dias IE, Castelo-Branco R, Scharfenstein H, Rezende de Castro R, Silva A, Schneider MPC, Araújo MJ, Martins R, Domingues VF, Nogueira F, Camões V, Vasconcelos VM, Leão PN (2019) Structure of hierridin C, synthesis of hierridins B and C, and evidence for prevalent alkylresorcinol biosynthesis in picocyanobacteria. J Nat Prod 82(2):393–402. https://doi.org/10.1021/acs.jnatprod.8b01038
Dedysh SN, Kulichevskaya IS, Beletsky AV, Ivanova AA, Rijpstra WIC, Damsté JSS, Mardanov AV, Ravin NV (2020) Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst Appl Microbiol 43(1):126050. https://doi.org/10.1016/j.syapm.2019.126050
Donner J, Reck M, Bunk B, Jarek M, App CB, Meier-Kolthoff JP, Overmann J, Müller R, Kirschning A, Wagner-Döbler I (2017) The biofilm inhibitor carolacton enters gram-negative cells: studies using a TolC-deficient strain of Escherichia coli. mSphere 2(5). https://doi.org/10.1128/mSphereDirect.00375-17
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL, USA
Eggeling L, Bott M (2015) A giant market and a powerful metabolism: L-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99(8):3387–3394. https://doi.org/10.1007/s00253-015-6508-2
Funabashi M, Funa N, Horinouchi S (2008) Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus*. J Biol Chem 283(20):13983–13991. https://doi.org/10.1074/jbc.M710461200
Gande R, Dover LG, Krumbach K, Besra GS, Sahm H, Oikawa T, Eggeling L (2007) The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J Bacteriol 189(14):5257–5264
Gonzalez AG, Bermejo Barrera J, Rodriguez Perez ME (1992) Synthesis of hierridin, a phenol from the lichen Ramalina hierrensis. Phytochemistry 31(4):1436–1439. https://doi.org/10.1016/0031-9422(92)80314-5
Hayashi T, Kitamura Y, Funa N, Ohnishi Y, Horinouchi S (2011) Fatty acyl-AMP ligase involvement in the production of alkylresorcylic acid by a Myxococcus xanthus type III polyketide synthase. ChemBioChem 12(14):2166–2176. https://doi.org/10.1002/cbic.201100344
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF (2016) A new view of the tree of life. Nat Microbiol 1(5):16048. https://doi.org/10.1038/nmicrobiol.2016.48
Ivanova A, Dedysh S (2012) Abundance, diversity, and depth distribution of Planctomycetes in acidic northern wetlands. Front Microbiol 3(5). https://doi.org/10.3389/fmicb.2012.00005
Kallscheuer N, Jogler C (2021) The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol Adv 53:107818. https://doi.org/10.1016/j.biotechadv.2021.107818
Kallscheuer N, Marienhagen J (2018) Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microbial Cell Fact 17(1):70. https://doi.org/10.1186/s12934-018-0923-x
Kallscheuer N, Vogt M, Stenzel A, Gätgens J, Bott M, Marienhagen J (2016) Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab Eng 38:47–55
Kallscheuer N, Vogt M, Bott M, Marienhagen J (2017) Functional expression of plant-derived O-methyltransferase, flavanone 3-hydroxylase, and flavonol synthase in Corynebacterium glutamicum for production of pterostilbene, kaempferol, and quercetin. J Biotechnol 258:190–196. https://doi.org/10.1016/j.jbiotec.2017.01.006
Kallscheuer N, Kage H, Milke L, Nett M, Marienhagen J (2019a) Microbial synthesis of the type I polyketide 6-methylsalicylate with Corynebacterium glutamicum. Appl Microbiol Biotechnol 103(23):9619–9631. https://doi.org/10.1007/s00253-019-10121-9
Kallscheuer N, Moreira C, Airs R, Llewellyn CA, Wiegand S, Jogler C, Lage OM (2019b) Pink- and orange-pigmented Planctomycetes produce saproxanthin-type carotenoids including a rare C45 carotenoid. Environ Microbiol Rep 11(6):741–748. https://doi.org/10.1111/1758-2229.12796
Kallscheuer N, Jeske O, Sandargo B, Boedeker C, Wiegand S, Bartling P, Jogler M, Rohde M, Petersen J, Medema MH, Surup F, Jogler C (2020) The planctomycete Stieleria maiorica Mal15T employs stieleriacines to alter the species composition in marine biofilms. Commun Biol 3(1):303. https://doi.org/10.1038/s42003-020-0993-2
Kallscheuer N, Rast P, Jogler M, Wiegand S, Kohn T, Boedeker C, Jeske O, Heuer A, Quast C, Glöckner FO, Rohde M, Jogler C (2021) Analysis of bacterial communities in a municipal duck pond during a phytoplankton bloom and isolation of Anatilimnocola aggregata gen. nov., sp. nov., Lacipirellula limnantheis sp. nov. and Urbifossiella limnaea gen. nov., sp. nov. belonging to the phylum Planctomycetes. Environ Microbiol 23(3):1379–1396. https://doi.org/10.1111/1462-2920.15341
Keilhauer C, Eggeling L, Sahm H (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175(17):5595–5603
Kogure T, Inui M (2018) Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl Microbiol Biotechnol 102(20):8685–8705. https://doi.org/10.1007/s00253-018-9289-6
Kortmann M, Kuhl V, Klaffl S, Bott M (2015) A chromosomally encoded T7 RNA polymerase-dependent gene expression system for Corynebacterium glutamicum: construction and comparative evaluation at the single-cell level. Microbial Biotechnol 8(2):253–265
Kulichevskaya IS, Ivanova AA, Baulina OI, Rijpstra WIC, Sinninghe Damsté JS, Dedysh SN (2017) Fimbriiglobus ruber gen. nov., sp. nov., a Gemmata-like planctomycete from Sphagnum peat bog and the proposal of Gemmataceae fam. nov. Int J Syst Evol Microbiol 67(2):218–224. https://doi.org/10.1099/ijsem.0.001598
Lage O, Bondoso J (2012) Bringing Planctomycetes into pure culture. Front Microbiol 3:405. https://doi.org/10.3389/fmicb.2012.00405
Lage OM, Bondoso J (2014) Planctomycetes and macroalgae, a striking association. Front Microbiol 5:267. https://doi.org/10.3389/fmicb.2014.00267
Leão PN, Costa M, Ramos V, Pereira AR, Fernandes VC, Domingues VF, Gerwick WH, Vasconcelos VM, Martins R (2013) Antitumor activity of hierridin B, a cyanobacterial secondary metabolite found in both filamentous and unicellular marine strains. PLoS ONE 8(7):e69562. https://doi.org/10.1371/journal.pone.0069562
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296. https://doi.org/10.1093/nar/gkab301
Lim CG, Fowler ZL, Hueller T, Schaffer S, Koffas MAG (2011) High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol 77(10):3451–3460. https://doi.org/10.1128/AEM.02186-10
Liu H-W, Begley T (2020) Comprehensive natural products III. Elsevier, Amsterdam, Netherlands
Martins TP, Rouger C, Glasser NR, Freitas S, de Fraissinette NB, Balskus EP, Tasdemir D, Leão PN (2019) Chemistry, bioactivity and biosynthesis of cyanobacterial alkylresorcinols. Nat Prod Rep 36(10):1437–1461
Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA (2014) A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comp Biol 10(12):e1004016. https://doi.org/10.1371/journal.pcbi.1004016
Meslet-Cladière L, Delage L, Leroux CJ-J, Goulitquer S, Leblanc C, Creis E, Gall EA, Stiger-Pouvreau V, Czjzek M, Potin P (2013) Structure/function analysis of a type III polyketide synthase in the brown alga Ectocarpus siliculosus reveals a biochemical pathway in phlorotannin monomer biosynthesis. Plant Cell 25(8):3089–3103. https://doi.org/10.1105/tpc.113.111336
Milke L, Marienhagen J (2020) Engineering intracellular malonyl-CoA availability in microbial hosts and its impact on polyketide and fatty acid synthesis. Appl Microbiol Biotechnol 104(14):6057–6065. https://doi.org/10.1007/s00253-020-10643-7
Milke L, Aschenbrenner J, Marienhagen J, Kallscheuer N (2018) Production of plant-derived polyphenols in microorganisms: current state and perspectives. Appl Microbiol Biotechnol 102(4):1575–1585. https://doi.org/10.1007/s00253-018-8747-5
Milke L, Kallscheuer N, Kappelmann J, Marienhagen J (2019) Tailoring Corynebacterium glutamicum towards increased malonyl-CoA availability for efficient synthesis of the plant pentaketide noreugenin. Microbial Cell Fact 18(1):71. https://doi.org/10.1186/s12934-019-1117-x
Milke L, Mutz M, Marienhagen J (2020) Synthesis of the character impact compound raspberry ketone and additional flavoring phenylbutanoids of biotechnological interest with Corynebacterium glutamicum. Microbial Cell Fact 19(1):92. https://doi.org/10.1186/s12934-020-01351-y
Nivina A, Yuet KP, Hsu J, Khosla C (2019) Evolution and diversity of assembly-line polyketide synthases. Chem Rev 119(24):12524–12547. https://doi.org/10.1021/acs.chemrev.9b00525
Overmann J, Abt B, Sikorski J (2017) Present and future of culturing bacteria. Annu Rev Microbiol 71(1):711–730. https://doi.org/10.1146/annurev-micro-090816-093449
Paczia N, Nilgen A, Lehmann T, Gätgens J, Wiechert W, Noack S (2012) Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microbial Cell Fact 11(1):1–14
Pandith SA, Ramazan S, Khan MI, Reshi ZA, Shah MA (2019) Chalcone synthases (CHSs): the symbolic type III polyketide synthases. Planta 251(1):15. https://doi.org/10.1007/s00425-019-03307-y
Panter F, Garcia R, Thewes A, Zaburannyi N, Bunk B, Overmann J, Gutierrez MV, Krug D, Müller R (2019) Production of a dibrominated aromatic secondary metabolite by a planctomycete implies complex interaction with a macroalgal host. ACS Chem Biol 14(12):2713–2719. https://doi.org/10.1021/acschembio.9b00641
Papendorf O, König GM, Wright AD (1998) Hierridin B and 2,4-dimethoxy-6-heptadecyl-phenol, secondary metabolites from the cyanobacterium Phormidium ectocarpi with antiplasmodial activity. Phytochemistry 49(8):2383–2386. https://doi.org/10.1016/S0031-9422(98)00440-3
Parage C, Tavares R, Réty S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais GAB, Aubourg S, Hugueney P (2012) Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol 160(3):1407–1419. https://doi.org/10.1104/pp.112.202705
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490. https://doi.org/10.1371/journal.pone.0009490
Rivas-Marin E, Wiegand S, Kallscheuer N, Jogler M, Peeters SH, Heuer A, Jetten MSM, Boedeker C, Rohde M, Devos DP, Jogler C (2020) Thalassoglobus polymorphus sp. nov., a novel Planctomycete isolated close to a public beach of Mallorca Island. Antonie Van Leeuwenhoek 113(12):1915–1926. https://doi.org/10.1007/s10482-020-01437-y
Rozmer Z, Perjési P (2016) Naturally occurring chalcones and their biological activities. Phytochem Rev 15(1):87–120. https://doi.org/10.1007/s11101-014-9387-8
Sambrook J, Russell D (2001) Molecular cloning, vol. 1–3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sandargo B, Jeske O, Boedeker C, Wiegand S, Wennrich J-P, Kallscheuer N, Jogler M, Rohde M, Jogler C, Surup F (2020) Stieleriacines, N-acyl dehydrotyrosines from the marine Planctomycete Stieleria neptunia sp. nov. Front Microbiol 11:1408. https://doi.org/10.3389/fmicb.2020.01408
Santana-Molina C, Henriques V, Hornero-Méndez D, Devos DP, Rivas-Marin E (2022) The squalene route to C30 carotenoid biosynthesis and the origins of carotenoid biosynthetic pathways. Proc Natl Acad Sci USA 119(52):e2210081119. https://doi.org/10.1073/pnas.2210081119
Thompson JD, Gibson TJ, Higgins DG (2003) Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics 2.3.1–2.3.22 doi:https://doi.org/10.1002/0471250953.bi0203s00
Vitorino IR, Lage OM (2022) The Planctomycetia: an overview of the currently largest class within the phylum Planctomycetes. Antonie Van Leeuwenhoek 115(2):169–201. https://doi.org/10.1007/s10482-021-01699-0
Wiegand S, Jogler M, Jogler C (2018) On the maverick Planctomycetes. FEMS Microbiol Rev 42(6):739–760. https://doi.org/10.1093/femsre/fuy029
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2020) Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol 5(1):126–140. https://doi.org/10.1038/s41564-019-0588-1
Wolf S, Becker J, Tsuge Y, Kawaguchi H, Kondo A, Marienhagen J, Bott M, Wendisch Volker F, Wittmann C (2021) Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem 65(2):197–212. https://doi.org/10.1042/ebc20200134
Zabolotneva AA, Shatova OP, Sadova AA, Shestopalov AV, Roumiantsev SA (2022) An overview of alkylresorcinols biological properties and effects. J Nutr Metab 2022:4667607. https://doi.org/10.1155/2022/4667607
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Project-ID 239748522 – CRC 1127 ChemBioSys (projects A06 and A07). CB greatly acknowledges funding from the European Union’s Horizon 2020 research and innovation program (ERC Grant number: 802736, MORPHEUS).
Deutsche Forschungsgemeinschaft,239748522,Nicolai Kallscheuer,239748522,Christine Beemelmanns,HORIZON EUROPE European Research Council,802736,Christine Beemelmanns,802736,Sven Balluff
Author information
Authors and Affiliations
Contributions
LM and KLC constructed the plasmids and prepared the ethyl acetate extracts; MK and RZ performed the plate assays for antibiotic susceptibility and antimicrobial activity; JG and SB performed the chromatographic analysis; KK prepared the medium and performed the cultivation; CEW performed the phylogenetic analysis and prepared the phylogenetic trees; CB, CJ, and JM contributed to data analysis and text preparation; NK performed the genome mining, constructed the planctomycetal deletion mutant, wrote the first draft of the manuscript, and supervised the study.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Milke, L., Kabuu, M., Zschoche, R. et al. A type III polyketide synthase cluster in the phylum Planctomycetota is involved in alkylresorcinol biosynthesis. Appl Microbiol Biotechnol 108, 239 (2024). https://doi.org/10.1007/s00253-024-13065-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00253-024-13065-x