Abstract
Shewanella oneidensis is a gram-negative bacterium known for its unique respiratory capabilities, which allow it to utilize a wide range of electron acceptors, including solid substrates such as electrodes. For a future combination of chemical production and electro-fermentation, the goal of this study was to expand its product spectrum. S. oneidensis was metabolically engineered to optimize its glutamate production and to enable production of itaconic acid. By deleting the glutamate importer gltS for a reduced glutamate uptake and pckA/ptA to redirect the carbon flux towards the TCA cycle, a ∆3 mutant was created. In combination with the plasmid pG2 carrying the glutamate dehydrogenase gdhA and a specific glutamate exporter NCgl1221 A111V, a 72-fold increase in glutamate concentration compared to the wild type was achieved. Along with overexpression of gdhA and NCgl1221 A111V, the deletion of gltS and pckA/ptA as well as the deletion of all three genes (∆3) was examined for their impact on growth and lactate consumption. This showed that the redirection of the carbon flux towards the TCA cycle is possible. Furthermore, we were able to produce itaconic acid for the first time with a S. oneidensis strain. A titer of 7 mM was achieved after 48 h. This suggests that genetic optimization with an expression vector carrying a cis-aconitate decarboxylase (cadA) and a aconitate hydratase (acnB) along with the proven redirection of the carbon flux to the TCA cycle enabled the production of itaconic acid, a valuable platform chemical used in the production of a variety of products.
Key points
•Heterologous expression of gdhA and NCgl1221_A111V leads to higher glutamate production.
•Deletion of ackA/pta redirects carbon flux towards TCA cycle.
•Heterologous expression of cadA and acnB enables itaconic acid production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
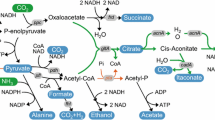
Glutamate and itaconic acid
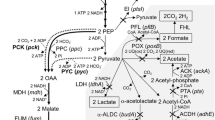
Natural amino acids like glutamate have a wide range of uses in the chemical and biotechnological industries. The most prevalent kind of glutamate in the food business is monosodium glutamate. In the pharmaceutical sector, glutamate is used to produce a variety of drugs, including antibiotics, anticancer drugs, and immunosuppressants (Ault 2004). The glutamic acid market is predicted to be valued USD 9.54 billion in 2020 and is expected to increase at a compound yearly growth rate of 7.6% from 2021 to 2028, reaching USD 17.16 billion. With a revenue share of more than 80% in 2020, the food and beverage application category led the glutamic acid market (Market Analysis Report 2022). Glutamate is mainly manufactured through microbial fermentation. Kinoshita et al. (2004) discovered the L-glutamate-producing bacterium, Corynebacterium glutamicum, originally designated Micrococcus glutamicus in 1957. Several bacteria applicable for glutamate production have been isolated since (Sano 2009). In general, commercially effective producers have been created by gradually accumulating advantageous genetic and phenotypic traits in a single background using traditional mutagenesis and/or recombinant DNA technologies. Making use of those technologies, a combination of gene deletion and plasmid based heterologous expression seemed applicable. In this study, the aim was to redirect the carbon flux from acetate towards the TCA cycle for an enhanced glutamate production and to reduce the reuptake of glutamate by host cells. Therefore, genes like pckA/ptA of Shewanella oneidensis MR-1 coding for enzymes catalyzing the ATP-generating steps from acetyl-CoA to acetate (Hunt et al. 2010) were targeted for deletion as well as a glutamate importer (gltS) to reduce the reuptake of secreted glutamate. To further optimize the glutamate production, an inducible vector pJem1 (obtained from (Jeske and Altenbuchner 2010) was constructed with the glutamate dehydrogenase gdhA and the specific glutamate exporter NCgl1221 A111V from Corynebacterium glutamicum (Nakamura et al. 2007). Such advances entail strains like S. oneidensis that are able to produce glutamate at better yields as well as fewer byproducts, as their removal accounts for the majority of expenses during downstream processing (Zhang et al. 2014; Kogure and Inui 2018).
Itaconic acid (IA) is a flexible platform substance that may be utilized as a monomer to create a variety of polymers, including biodegradable plastics (Sriariyanun et al. 2019; Ray et al. 2017). Asia Pacific accounts for about 54% of the worldwide demand for itaconic acid, with Europe and North America coming in second and third. Methyl methacrylate, polyitaconic acid, and SBR (styrene-butadiene rubber) latex manufacture account for a sizeable percentage of the itaconic acid market. The global market was worth around $75 million in 2015, growing to $95.4 million by 2021, and is predicted to be worth around $110.4 million by 2028 (Global Market Insights 2016; Market Data Forecast 2023). Aspergillus itaconicus was shown by (Kinoshita 1932) to create IA when grown on D-glucose, providing the first instance of a microbe capable of doing so. The Northern Regional Research Laboratory (NRRL) of the United States Department of Agriculture discovered Aspergillus terreus as a unique producer of IA in 1939 and identified A. terreus NRRL 1960 as a strain able to generate large titers of IA (80 g L−1) (Lockwood and Nelson 1946). Because of A. terreus’ extreme sensitivity to contaminants (e.g., manganese) in the fermentation broth (Karaffa et al. 2015) and the morphological diversity (in particular as pellets of different sizes or filamentous) in which this fungus can grow (Krull et al. 2017), its utilization for IA synthesis in an industrial context is challenging, pointing out the need for new production hosts. This study approaches to modify S. oneidensis genetically to produce IA from its naturally produced intermediate cis-aconitate. The same steps as for the enhanced glutamate production were taken here to redirect the carbon flux towards the TCA cycle. The missing enzyme for IA production, a cis-aconitate decarboxylase from A. terreus itself (cadA), will be heterologously expressed on the inducible vector pJem1 (obtained from (Jeske and Altenbuchner 2010). To increase the production of the precursor cis-aconitate and consequently IA production, it would be reasonable to overexpress S. oneidensis acnB, which encodes the aconitate hydratase.
Shewanella oneidensis and bioelectrotechnology
S. oneidensis is a gram-negative bacterium known for its unique respiratory capabilities that allow it to utilize a variety of electron acceptors, including solid substrates such as electrodes (Marsili et al. 2008). Due to this characteristic, S. oneidensis has become a popular choice for the realization of microbial electrosynthesis (MES) and microbial fuel cells (MFC), which depend on the transmission of electrons from microorganisms to an electrode to produce energy or chemicals (Ikeda et al. 2021). Schröder et al. (2015) define MES as “the execution of microbially catalyzed electrochemical reactions to transform a substance into a desired product.” Electroactive bacteria (EAB) can transfer electrons across biological membranes to connect intracellular and external electron acceptor/donor systems. These EAB may be utilized in either MFC or MES. Bacteria in MFCs oxidize organic fuels such as lactate as well as complex combinations of organic matter and transfer the biologically produced electrons to the anode, resulting in current generation. Depending on the intended reaction, MES can be classified as cathodic or anodic. Bacteria in cathodic MES receive electrons given by external sources (e.g., electrodes) and utilize them in reduction processes (e.g., reductive carbon fixation (Rabaey et al. 2009)). In anodic MES, bacteria give off electrons during an unbalanced fermentation (Flynn et al. 2010). MES applications for S. oneidensis include, for instance, bioremediation of dyes (Gomaa et al. 2017; Li et al. 2018b), metal ions (Han et al. 2017), nitrobenzene (Wang et al. 2013), and sulfonamides (Mao et al. 2018).
Extracellular electron transfer is characterized in three ways: direct electron transfer (DET), mediated electron transfer (MET), and indirect electron transfer (IET). DET necessitates physical contact with the electrode, which may be achieved by the use of pili, nanowires, or membrane-bound cytochromes. It is commonly used by bacteria that create biofilms, such as Geobacter sulfurreducens. No physical contact is required for the MET; instead, bacteria such as S. oneidensis employ redox active mediator molecules such as flavines to transport electrons between the cell and the electrode. These mediator molecules can be regenerated electrophilically unlike electron-shuttling chemicals needed for IET like hydrogen or formic acid which are consumed by microorganisms such as C. necator (Sydow et al. 2014).
The aim of this study was to broaden the product spectrum of S. oneidensis by metabolic engineering for a future combination of chemical production with electro-fermentation. So far, the production of isobutanol (Jeon et al. 2015), acetoin (Bursac et al. 2017), and formic acid (Le Tuan et al. 2018) could be shown with S. oneidensis.
Materials and methods
Chemicals
All chemicals were obtained from Carl Roth (Karlsruhe, Germany), VWR International (Darmstadt, Germany), or Merck (Darmstadt, Germany). Solvents for chromatography were acquired of LC–MS grade quality.
Bacterial strains and growth conditions
Escherichia coli DH5α (ATCC 67879 (Hanahan 1985; Grant et al. 1990)) and E. coli WM3064 cultures were grown in Lysogeny broth (LB) medium (Bertani 1951) at 37 °C. E. coli WM3064 cultures were supplemented with 0.3 mM of 2,6-diaminopimelic acid (DAP) and supplied with 50 μg/mL of kanamycin sulfate when necessary. For cultivation of S. oneidensis MR-1 (ATCC 700550 (Myers and Nealson 1988)), lactate Shewanella basal medium (LSBM) was prepared containing the following compounds per liter: KH2PO4 225 mg, K2HPO4 225 mg, NaCl 460 mg, (NH4)2SO4 225 mg, MgSO4 × 7 H2O 117 mg, HEPES 23.8 g, lactate (90% solution) 22.4 g (100 mM), 5 mL vitamin solution, and 5 mL trace mineral solution. The pH was adjusted to 7.2 before adding the vitamin and trace mineral solution using a 1 M NaOH solution made from NaOH pellets. The vitamin solution contained per liter biotin 2 mg, folic acid 2 mg, pyridoxine hydrochloride 10 mg, thiamin hydrochloride 5 mg, riboflavin 5 mg, nicotinic acid 5 mg, DL-calcium pantothenate 5 mg, cyanocobalamine 0.1 mg, 4-aminobenzoic acid 5 mg, and lipoic acid 5 mg. The trace mineral solution contained the following ingredients per liter: nitrilotriacetic acid (C6H9NO6) 1.5 g, MgSO4 × 7 H2O 3 g, MnSO4 × 2 H2O 0.5 g, NaCl 1 g, FeSO4 × 7 H2O 100 mg, CoCl2 100 g, CaCl2 × 2 H2O 100 mg, ZnCl2 130 mg, CuSO4 × 5 H2O 10 mg, AlK(SO4)2 10 mg, H3BO3 10 mg, Na2MoO4 × 2 H2O 25 mg, NiCl2 24 mg, and Na2WO4 × 2 H2O 25 mg. The LSBM main solution was sterilized for 20 min at 121 °C, and the vitamin and trace mineral solutions were sterile filtered and kept at 4 °C until further use. Five milliliter precultures in LB medium were cultured in test tubes for 24 h at 30 °C and 180 rpm in an orbital shaking incubator (Minitron HT, Infors, Bottmingen, Switzerland) for growth of S. oneidensis MR-1 in liquid media. Main cultures of 25 mL in LSBM supplemented with 0.1% (wt/vol) casein hydrolysate were subsequently inoculated to an OD600 of 0.05 in 100 mL shake flasks and incubated under the same conditions. If required, kanamycin sulfate at 50 μg/mL was added to the medium. For promoter induction from plasmid pJeM1 and pG2, l-rhamnose was added to a final concentration of 0.2% (wt/vol) at OD600 of 0.15–0.3 in liquid medium if not stated otherwise.
Cultivation of S. oneidensis MR-1 in BioLector microbioreactor and fluorescence assay
The precultures of S. oneidensis MR-1 were grown in LB for 16 h and used for the inoculation of 1 mL medium at the starting OD600 of 0.05. The cultivation was carried out in a BioLector® MB system (m2p-labs, Germany) in MTP-48 FlowerPlates® with pH optodes at 30 °C, 1000 rpm, and 95% humidity. The growth of the cultures was tracked online using scattered light signal monitoring. To examine the induction of the rhaBAD promoter by different concentrations of l-rhamnose and its influence on bacterial growth, fluorescence of bacterial suspension as well as the optical density was continuously recorded. The fluorescence intensity at 488 nm excitation wavelength and 520 nm emission wavelength was measured and normalized by the optical density at 600 nm.
DNA cloning and plasmid construction
All plasmid cloning techniques were carried out in E. coli DH5α. Thermo Scientific’s GeneJET Plasmid Miniprep Kit (Waltham, USA) was used to purify plasmid DNA. Polymerase chain reactions (PCR) were carried out according to the manufacturer’s procedure using Q5 Polymerase from New England Biolabs (Frankfurt, Germany). Following that, PCR products were purified using the Zymo Research Europe (Freiburg, Germany) DNA Clean & Concentrator Kit. Vectors were assembled using the isothermal in vitro recombination (Gibson et al. 2009). Oligonucleotides were purchased from Merck (Darmstadt, Germany) and restriction enzymes and T4 ligase from NEB. Sanger sequencing at Eurofins Scientific (Luxembourg, Luxembourg) verified all genetic constructs. Transformation of E. coli DH5α and WM3064 was done via electroporation. Vectors were conjugated in S. oneidensis MR-1 with E. coli WM3064 as donor strain. Genomic DNA of S. oneidensis MR-1 was purified with GenElute™ Bacterial Genomic DNA Kit from Merck (Darmstadt, Germany). Oligonucleotides, plasmids, and strains used in this work can be found in Table S1 in the supplementary information.
Generation of deletion mutants
Knockout of the genes ackA (SO 2915), pta (SO 2916), and gltS (SO3562) was carried out with allelic exchange vector pNTPS138-R6KT as described by Lassak et al. (2010). Upstream and downstream portions (approximately 500 bp) of the targeted gene area, as well as the backbone vector pNTPS138-R6KT, were amplified by PCR using the relevant primer pairs (given in Table S1 in the supplementary information). Gibson Assembly fused the fragments after purification.
Metabolite analysis
Metabolites were quantified by HPLC equipped with a UV–visible-light (Vis) detector and refractive index detector. The HPLC system consisted of an SPD-20A UV–Vis detector, two LC-20AT pumps, a SIL-20AC autosampler, a CTO-20AC column oven, and an RID-10A detector. The units used are from the manufacturer Shimadzu Deutschland GmbH (Duisburg, Germany). A Rezex™ ROA-Organic Acid H + 8% column (250 × 4.6 mm; Phenomenex, California, USA) with precolumn was used for the analysis. Flow rate was 0.6 mL/min, the column oven temperature 30 °C, injection volume 10 μL, and the separation time was 35 min. The mobile phase consisted of acidified water (0.005 M H2SO4). For quantification of metabolites, the supernatant of S. oneidensis cultures was centrifuged for 5 min at 16.000 g and passed through a 0.22 μm PVDF-syringe filter (Carl Roth, Karlsruhe, Germany). Samples were analyzed at 254 nm for lactate and 202 nm for itaconic acid and acetate. Retention times of the analytes are listed in Table S2 in the supplemental material. For the quantification of glutamate, a LC–MS/MS setup was used. One microliter of each sample was chromatographically purified on a 150 × 4.6 mm Luna Omega 3 m PS C18 100 column (Phenomenex, Aschaffenburg, Germany) in a Nexera X2 UHPLC system (Shimadzu, Duisburg, Germany). The mobile phase consisted of 0.2% formic acid in water (A) and 0.2% formic acid in acetonitrile (B). The pump gradient started at 10% mobile phase B; from 0.10 min, the proportion of mobile phase B was continuously increased linearly until it reached 100% B at 2 min; from 2.50 min to 4.50 min, the proportion of mobile phase B was again decreased linearly to the initial value of 10%. The total run time was 5 min. The flow rate was 0.5 mL/min. The analytes were ionized negatively with an APCI ion source, fragmented, and ultimately quantified by comparing the findings to calibration curves of comparable standards. Shimadzu’s LabSolutions software was used for quantification.
Software
Plasmid design was performed using SnapGene® software (from Dotmatics; available at snapgene.com). All graphs were generated using Origin(Pro), version 2022b (OriginLab Corporation, Northampton, MA, USA).
GenBank accession numbers (Table 1 )
Results
Heterologous expression of gdhA and NCgl1221
S. oneidensis can use fermentation end products like lactate, formate, and hydrogen as electron donors and is capable of using a broad range of electron acceptors (e.g., fumarate, metal ions). This makes it an attractive host for biotechnological applications. The production of glutamate was studied as a model reaction to see if it is possible to modify S. oneidensis’ citrate cycle for the production of bulk chemicals. For this, the genes from Corynebacterium glutamicum for a glutamate dehydrogenase gdhA as well as the mutant of a mechano-selective glutamate exporter NCgl1221 A111V (Nakamura et al. 2007) were expressed from a pJeM1 plasmid (obtained from (Jeske and Altenbuchner 2010)) under control of the rhaBAD operon. As there was no published data about the use of this operon in S. oneidensis at the time of these experiments, its expression strength was measured using pJeM1 with GFP (green fluorescent protein) as reporter. Different rhamnose concentrations (0.002, 0.02, and 0.2%) were tested to observe the inducer’s impact on S. oneidensis’ growth (Fig. 1A) and to assess the promoter’s regulation measured via fluorescence (Fig. 1B). Later, Cheng and colleagues engineered a rhamnose-inducible system for various Shewanella species, which confirms our results (Cheng et al. 2021).
Influence of different L-rhamnose concentrations on a S. oneidensis’ growth and b relative fluorescence (0.2% rhamnose was set as 100%). The cultivations and measurements were performed in BioLector microbioreactor; the data points are the averages and standard deviations of three biological replicates. GFP fluorescence was measured at 520 nm
Higher concentrations of rhamnose led to an increase in relative fluorescence and had no impact on S. oneidensis growth. As this shows that the rhaBAD operon is suitable for S. oneidensis, the following experiments were conducted with the above-mentioned plasmid system, further referred to as pG2. The glutamate production of the wild-type strain carrying pG2 was compared to genetically optimized strains later on (Fig. 4).
Optimization of glutamate production by gene deletions
In addition to overexpression, several gene deletions were screened for their influence on glutamate production. S. oneidensis metabolizes lactate via pyruvate and acetyl-CoA to acetate using lactate as an electron sink if the amount of electron acceptor is limited (Takuya Kasai et al. 2019). The ackA/pta genes, which catalyze the ATP-generating steps from acetyl-CoA to acetate, were deleted to reroute the carbon flux from acetate to the TCA cycle (Hunt et al. 2010). S. oneidensis’ growth under aerobic conditions was not impacted by the deletions and is comparable to the wild type (Fig. 2a). In the deletion mutant ∆ackaA∆pta (∆2), generated with the pNPTS138-R6KT_ackApta deletion plasmid, acetate production (Fig. 2b) decreased significantly and remained at around 2 mM, probably due to background activity, while acetate production reached around 14 mM in the wild type after 9 h. After 24 h, a decrease in acetate concentration is observed in the wild type as it is taken up and metabolized again.
For further optimization of the glutamate production, reuptake of the produced glutamate must be prevented. Therefore, the gene gltS which codes for a glutamate transporter was deleted with the deletion plasmid pNPTS138-R6KT_gltS.
The wild-type and deletion mutants were grown with the empty plasmid pJeM1 as well as the generated plasmid pG2 to determine the gene deletion’s and plasmid’s influence on growth and substrate consumption. Under aerobic conditions, neither appeared to affect S. oneidensis’ growth (Fig. 3a) or lactate consumption (Fig. 3b). The slight differences in OD600nm are due to slightly varying initial values, but the growth curves follow the same pattern.
Influence of gene deletions and plasmids on a S. oneidensis’ growth and b lactate consumption. Δ3: mutant with deletion of ackA/pta and gltS; pJeM1: empty plasmid; pG2: plasmid containing glutamate production genes. The data points are the averages and standard deviations of three biological replicates
Comparing the influence of the empty plasmid (pJeM1) with plasmid pG2, a significantly higher glutamate production was evident for the strains with the latter, even though no clear overproduction of the proteins GdhA and NCgl1221 was evident on the SDS PAGE gels (data not shown). The glutamate concentrations in the strains carrying the empty plasmid remained below 1 mM (Fig. 4.).
The Δ3 deletion mutant showed the highest glutamate concentration with 25 mM, while that of the ΔgltS mutant was just under 17 mM. The glutamate concentration of the ΔackApta mutant was approximately 7 mM, and that of the wild type reached just under 5 mM, decreasing again after 22 h. It seems that the deletion of the glutamate exporter has the highest impact on the glutamate concentration.
Since one glutamate molecule is produced from three lactate molecules (Eq. 1), it can be deduced that with the Δ3 deletion mutant nearly 76% of the theoretical maximum of the consumed lactate was converted into glutamate. Considering the differences in fermentation, especially the lower biomass for S. oneidensis, the biomass-specific glutamate production is in a comparable range to that of industrially used C. glutamicum strains (Table 2). The final product concentrations in the newly developed system are even lower by a factor of 27. Therefore, it can be concluded that the next step here is to optimize the growth of S. oneidensis.
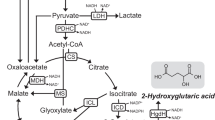
Production of itaconic acid
As the metabolic engineering to increase glutamate production was successful, the next step was to broaden the product spectrum of S. oneidensis. In order to produce itaconic acid from cis-aconitate, which is a natural intermediate in the citrate cycle of S. oneidensis, the gene for a cis-aconitate decarboxylase (cadA) from Aspergillus terreus must be heterologously expressed. In addition, S. oneidensis’ gene for the aconitate hydratase (acnB) needs to be overexpressed to achieve a higher production of the precursor cis-aconitate.
According to the glutamate production, the above-mentioned genes (cadA and acnB) were expressed from a pJeM1 plasmid (obtained from (Jeske and Altenbuchner 2010)) under control of the rhaBAD operon, further referred to as plasmid pIA. For the wild type carrying the pIA plasmid, the itaconic acid production remained at around 0.5 mM after 72 h, while the ∆ackA/pta mutant with pIA reached titers of nearly 7 mM after 48 h (Fig. 5). Without the plasmid pIA, no itaconic acid production was observed.
Even though the product titers and yields for itaconic acid are low compared to the industrial used strains like A. terreus DSM 23081, this is the first reported S. oneidensis strain producing itaconic acid (Table 3).
Discussion
Metabolic engineering is widely used to develop strains with enhanced abilities for a variety of applications. As S. oneidensis MR-1 is a model electroactive bacterium for MFCs, most studies focus on engineering strategies for the optimization of EET (Li et al. 2022, 2018a), broadening the substrate spectrum (Li et al. 2017; Sekar et al. 2016; Choi et al. 2014), and biofilm formation (Mukherjee et al. 2020). In this work, we focused on expanding the product spectrum by redirecting the carbon flux of the TCA cycle. Due to its electroactive characteristics and its genetic accessibility, S. oneidensis is a promising host for the coupling of bulk chemical production with MES and MFC applications. As proof of concept, we optimized its glutamate production and successfully achieved a 72-fold increase in glutamate concentration with our deletion mutant ∆3 carrying the plasmid pG2 compared to the wild type. The overexpression of a glutamate dehydrogenase and the glutamate exporter had the highest impact with a 13-fold rise of glutamate, due to the increased abundance of the enzymes that are involved in glutamate synthesis and export. The deletion of the glutamate importer (∆gltS) led to a fivefold increase by limiting the reuptake of glutamate and complete secretion into the medium. This emphasizes the power of overexpression of the glutamate dehydrogenase and exporter as only by the blocked reuptake of glutamate the enormous increase of glutamate in the medium is measurable. Another 1.5-fold surge was achieved by deleting the acetate-producing genes as its production no longer competed with the carbon source. To our knowledge, this is the first optimized Shewanella strain to produce glutamate at this scale. Furthermore, in this work, we were able to produce itaconic acid for the first time with a S. oneidensis strain. Deletion of the acetate-producing genes led to an almost 13-fold increase in itaconic acid production. To further optimize its production via metabolic engineering, the mfsA gene, which encodes a membrane permease responsible for the secretion of the produced itaconic acid into the extracellular environment, could also be overexpressed (Huang et al. 2014). Other options include deletion of the malate dehydrogenase gene and downregulation of isocitrate dehydrogenase as shown by (Harder et al. 2016) for E. coli.
Glutamate and itaconic acid production both lead to the production of excess electrons and thus to unwanted by-products to maintain the redox balance. Electroactive bacteria, in turn, can transfer these excess electrons to an external electrode, removing the need to produce undesirable side products and enabling stoichiometric conversion of substrate to product while also producing electrical current. This was shown by (Flynn et al. 2010) with an engineered S. oneidensis strain, which could convert glycerol to ethanol only in the presence of an oxidizing electrode. One of the many potential advantages of electrode-linked microbial catalysis is that it can be used as an electrochemical “lever” to drive an unfavorable reaction, enable the operation of a microbial fuel cell to generate electricity, or act as reducing equivalents for the synthesis of additional products (Rabaey and Rozendal 2010). (Bursac et al. 2017) developed a S. oneidensis strain which produced acetoin with a yield of 78% of the theoretical acetoin production maximum by electro-fermentation. Therefore, to increase the titers of the strains generated in this work, the next step would be to cultivate them in a BES with an oxidizing electrode. Future research will need to demonstrate whether it is possible to combine the two research fields - broadening the range of substrates and employing unbalanced fermentation - to achieve productivities that permit a competitive bio(electro)technological production using S. oneidensis as the producing strain.
In conclusion, our work effectively used metabolic engineering to boost S. oneidensis’ production capacities. When compared to the wild type, the genetically engineered strain produced up to 25 mM glutamate, a remarkable 72-fold increase. Furthermore, for the first time, we showed that a genetically engineered S. oneidensis strain could produce itaconic acid. These findings support that it is possible to redirect carbon flux to the tricarboxylic acid (TCA) cycle in S. oneidensis. These findings enable future efforts to combine chemical production with electro-fermentation, a promising approach for sustainable and energy-efficient processes.
References
Abdenacer M, Aicha N, Joseph B, Nabil N (2021) High production of L-glutamic acid from date juice extracts by Corynebacterium glutamicum using fed-batch cultures: pulsed and continuous feeding modes. ASN 8:37–50. https://doi.org/10.2478/asn-2021-0004
Ault A (2004) The monosodium glutamate story: the commercial production of MSG and other amino acids. J Chem Educ 81:347. https://doi.org/10.1021/ed081p347
Bursac T, Gralnick JA, Gescher J (2017) Acetoin production via unbalanced fermentation in Shewanella oneidensis. Biotechnol Bioeng 114:1283–1289. https://doi.org/10.1002/bit.26243
Cheng L, He R-L, Min Di, Li W-W, Liu D-F, Yu H-Q (2021) Engineering a rhamnose-inducible system to enhance the extracellular electron transfer ability of Shewanella genus for improved Cr(VI) reduction. ACS ES&T Eng 1:842–850. https://doi.org/10.1021/acsestengg.0c00125
Choi D, Lee SB, Kim S, Min B, Choi I-G, Chang IS (2014) Metabolically engineered glucose-utilizing Shewanella strains under anaerobic conditions. Bioresour Technol 154:59–66. https://doi.org/10.1016/j.biortech.2013.12.025
Flynn JM, Ross DE, Hunt KA, Bond DR, Gralnick JA (2010) Enabling unbalanced fermentations by using engineered electrode- interfaced bacteria. Mbio 1. https://doi.org/10.1128/mBio.00190-10
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. https://doi.org/10.1038/nmeth.1318
Global Market Insights (2016) Itaconic acid market. https://www.gminsights.com/industry-analysis/itaconic-acid-market. Accessed 26 April 2023
Gomaa OM, Fapetu S, Kyazze G, Keshavarz T (2017) The role of riboflavin in decolourisation of Congo red and bioelectricity production using Shewanella oneidensis-MR1 under MFC and non-MFC conditions. World J Microbiol Biotechnol 33:56. https://doi.org/10.1007/s11274-017-2223-8
Han J-C, Chen G-J, Qin L-P, Mu Y (2017) Metal respiratory pathway-independent Cr isotope fractionation during Cr (VI) reduction by Shewanella oneidensis MR-1. Environ Sci Technol Lett 4:500–504
Harder B-J, Bettenbrock K, Klamt S (2016) Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab Eng 38:29–37. https://doi.org/10.1016/j.ymben.2016.05.008
Harder B-J, Bettenbrock K, Klamt S (2018) Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol Bioeng 115:156–164. https://doi.org/10.1002/bit.26446
Huang X, Lu X, Li Y, Li X, Li J-J (2014) Improving itaconic acid production through genetic engineering of an industrial Aspergillus terreus strain. Microb Cell Fact 13:119. https://doi.org/10.1186/s12934-014-0119-y
Hunt KA, Flynn JM, Naranjo B, Shikhare ID, Gralnick JA (2010) Substrate-level phosphorylation is the primary source of energy conservation during anaerobic respiration of Shewanella oneidensis strain MR-1. J Bacteriol 192:3345–3351. https://doi.org/10.1128/JB.00090-10
Ikeda S, Takamatsu Y, Tsuchiya M, Suga K, Tanaka Y, Kouzuma A, Watanabe K (2021) Shewanella oneidensis MR-1 as a bacterial platform for electro-biotechnology. Essays Biochem 65:355–364. https://doi.org/10.1042/EBC20200178
Jeon J-M, Park H, Seo H-M, Kim J-H, Bhatia SK, Sathiyanarayanan G, Song H-S, Park S-H, Choi K-Y, Sang B-I, Yang Y-H (2015) Isobutanol production from an engineered Shewanella oneidensis MR-1. Bioprocess Biosyst Eng 38:2147–2154. https://doi.org/10.1007/s00449-015-1454-z
Jeske M, Altenbuchner J (2010) The Escherichia coli rhamnose promoter rhaP(BAD) is in Pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl Microbiol Biotechnol 85:1923–1933. https://doi.org/10.1007/s00253-009-2245-8
Karaffa L, Diaz R, Papp B, Fekete E, Sándor E, Kubicek CP (2015) A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl Microbiol Biotechnol 99:7937–7944
Kasai T, Suzuki Y, Kouzuma A, Watanabe K (2019) Roles of d-lactate dehydrogenases in the anaerobic growth of Shewanella oneidensis MR-1 on sugars. Appl Environ Microbiol 85(3):e02668-18. https://doi.org/10.1128/AEM.02668-18
Kinoshita K (1932) Über die Produktion von Itaconsäure und Mannit durch einen neuen Schimmelpilz Aspergillus itaconicus. Acta Phytochim 5:271–287
Kinoshita S, Udaka S, Shimono M (2004) Studies on the amino acid fermentation. Part 1. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol 50:331–343
Kogure T, Inui M (2018) Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl Microbiol Biotechnol 102:8685–8705. https://doi.org/10.1007/s00253-018-9289-6
Krull S, Hevekerl A, Kuenz A, Prüße U (2017) Process development of itaconic acid production by a natural wild type strain of Aspergillus terreus to reach industrially relevant final titers. Appl Microbiol Biotechnol 101:4063–4072
Kuenz A, Gallenmüller Y, Willke T, Vorlop K-D (2012) Microbial production of itaconic acid: developing a stable platform for high product concentrations. Appl Microbiol Biotechnol 96:1209–1216. https://doi.org/10.1007/s00253-012-4221-y
Le Tuan QA, Kim HG, Kim YH (2018) Electrochemical synthesis of formic acid from CO2 catalyzed by Shewanella oneidensis MR-1 whole-cell biocatalyst. Enzyme Microb Technol 116:1–5. https://doi.org/10.1016/j.enzmictec.2018.05.005
Li F, Li Y, Sun L, Li X, Yin C, An X, Chen X, Tian Y, Song H (2017) Engineering Shewanella oneidensis enables xylose-fed microbial fuel cell. Biotechnol Biofuels 10:196. https://doi.org/10.1186/s13068-017-0881-2
Li F, Li Y-X, Cao Y-X, Wang L, Liu C-G, Shi L, Song H (2018a) Modular engineering to increase intracellular NAD(H/+) promotes rate of extracellular electron transfer of Shewanella oneidensis. Nat Commun 9:3637. https://doi.org/10.1038/s41467-018-05995-8
Li Q, Feng X-L, Li T-T, Lu X-R, Liu Q-Y, Han X, Feng Y-J, Liu Z-Y, Zhang X-J, Xiao X (2018b) Anaerobic decolorization and detoxification of cationic red X-GRL by Shewanella oneidensis MR-1. Environ Technol 39:2382–2389. https://doi.org/10.1080/09593330.2017.1355933
Li Y, Li Y, Chen Y, Cheng M, Yu H, Song H, Cao Y (2022) Coupling riboflavin de novo biosynthesis and cytochrome expression for improving extracellular electron transfer efficiency in Shewanella oneidensis. Biotechnol Bioeng 119:2806–2818. https://doi.org/10.1002/bit.28172
Lockwood LB, Nelson GE (1946) Some factors affecting the production of itaconic acid by Aspergillus terreus in agitated cultures. Arch Biochem 10:365–374
Mao F, Liu X, Wu K, Zhou C, Si Y (2018) Biodegradation of sulfonamides by Shewanella oneidensis MR-1 and Shewanella sp. strain MR-4. Biodegradation 29:129–140. https://doi.org/10.1007/s10532-017-9818-5
Market Analysis Report (2022) Glutamic acid market size, share & trends analysis report by application (food & beverages, pharmaceuticals, animal feed), by region (North America, Europe, APAC, CSA, MEA), and segment forecasts, 2021–2028. https://www.grandviewresearch.com/industry-analysis/glutamic-acid-market
Market Data Forecast (2023) Itaconic acid market. https://www.marketdataforecast.com/market-reports/itaconic-acid-market. Accessed 26 April 2023
Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR (2008) Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci 105:3968–3973. https://doi.org/10.1073/pnas.0710525105
Mukherjee M, Zaiden N, Teng A, Hu Y, Cao B (2020) Shewanella biofilm development and engineering for environmental and bioenergy applications. Curr Opin Chem Biol 59:84–92. https://doi.org/10.1016/j.cbpa.2020.05.004
Nakamura J, Hirano S, Ito H, Wachi M (2007) Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce L-glutamic acid production. Appl Environ Microbiol 73:4491–4498. https://doi.org/10.1128/AEM.02446-06
Rabaey K, Angenent L, Schröder U, Keller J (2009) Bioelectrochemical systems: from extracellular electron transfer to biotechnological application. IWA Publishing. https://doi.org/10.2166/9781780401621
Rabaey K, Rozendal RA (2010) Microbial electrosynthesis - revisiting the electrical route for microbial production. Nat Rev Microbiol 8:706–716. https://doi.org/10.1038/nrmicro2422
Ray P, Smith C, Simon GP, Saito K (2017) Renewable green platform chemicals for polymers. Molecules 22(3):376. https://doi.org/10.3390/molecules22030376
Sano C (2009) History of glutamate production. Am J Clin Nutr 90:728S-732S. https://doi.org/10.3945/ajcn.2009.27462F
Schröder U, Harnisch F, Angenent LT (2015) Microbial electrochemistry and technology: terminology and classification. Energy Environ Sci 8:513–519. https://doi.org/10.1039/C4EE03359K
Sekar R, Shin HD, DiChristina TJ (2016) Activation of an otherwise silent xylose metabolic pathway in Shewanella oneidensis. Appl Environ Microbiol 82:3996–4005. https://doi.org/10.1128/AEM.00881-16
Sriariyanun M, Heitz JH, Yasurin P, Asavasanti S, Tantayotai P (2019) Itaconic acid: a promising and sustainable platform chemical? Appl Sci Eng Prog 12(2):75–82
Sydow A, Krieg T, Mayer F, Schrader J, Holtmann D (2014) Electroactive bacteria—molecular mechanisms and genetic tools. Springer Verlag, Eur J Appl Microbiol Biotechnol, p 98
Wang J, Lu H, Zhou Y, Song Y, Liu G, Feng Y (2013) Enhanced biotransformation of nitrobenzene by the synergies of Shewanella species and mediator-functionalized polyurethane foam. J Hazard Mater 252–253:227–232. https://doi.org/10.1016/j.jhazmat.2013.02.040
Zhang D, Guan D, Liang J, Guo C, Xie X, Zhang C, Xu Q, Chen N (2014) Reducing lactate secretion by ldhA deletion in L-glutamate- producing strain Corynebacterium glutamicum GDK-9. Braz J Microbiol 45:1477–1483. https://doi.org/10.1590/s1517-83822014000400044
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors gratefully acknowledge the financial support from the German Federal Ministry of Education and Research (031A226).
Author information
Authors and Affiliations
Contributions
HW and DH conceived and designed the experiment. HW, LvdS, and LZ conducted experiments. HW, LvdS, and DH drafted the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information file. The supplementary information file contains Table S1 with all oligonucleotides, plasmids, and strains used in this work and Table S2 with retention times of analytical standards used for the quantification of metabolites by S. oneidensis.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wohlers, H., Zentgraf, L., van der Sande, L. et al. Metabolic engineering of Shewanella oneidensis to produce glutamate and itaconic acid. Appl Microbiol Biotechnol 108, 36 (2024). https://doi.org/10.1007/s00253-023-12879-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00253-023-12879-5