Abstract
Pseudomonas aeruginosa is an emerging threat for hospitalized and cystic fibrosis patients. Biofilm, a microbial community embedded in extracellular polymeric substance, fortifies bacteria against the immune system. In biofilms, the expression of functional amyloids is linked with highly aggregative, multi-resistant strains, and chronic infections. Serrapeptase (SPT), a protease possessing similar or superior anti-microbial properties with many antibiotics, presents anti-amyloid potential. However, studies on the employment of SPT against Pseudomonas biofilms and Fap amyloid, or the possible mechanisms of action are scarce. Here, SPT inhibited biofilm formation of P. aeruginosa ATCC 27853 on both plastic and glass surfaces, with an IC50 of 11.26 µg/mL and 0.27 µg/mL, respectively. The inhibitory effect of SPT on biofilm was also verified with optical microscopy of crystal violet-stained biofilms and with confocal microscopy. Additionally, SPT caused a dose-dependent decrease of bacterial viability (IC50 of 3.07 µg/mL) as demonstrated by MTT assay. Reduction of bacterial functional amyloids was also demonstrated, employing both fluorescence microscopy with thioflavin T and photometrical determination of Congo-red-positive compounds. Both viability and functional amyloids correlated significantly with biofilm inhibition. Finally, in silico molecular docking studies provided a mechanistic insight into the interaction of SPT with FapC or FapD, proving that both peptides are possible targets of SPT. These results offer new insights into the biofilm formation of P. aeruginosa and potentiate the involvement of SPT in the prevention and eradication of Pseudomonas biofilms.
Graphical abstract

Key points
• Serrapeptase inhibits biofilm formation of P. aeruginosa on plastic and glass.
• Biofilm inhibition correlated with reduced viability and functional amyloid levels.
• In silico studies indicated that serrapeptase may target FapC and FapD peptides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas aeruginosa is a Gram-negative, non-fastidious, facultative anaerobic bacterial species, with low nutritional requirements. Thus, it can colonize most surfaces, with a preference for moist installations like hospital room sinks, toilets, showers, patient care equipment (respiratory ventilators, contact lenses, dialysis tubes), and even soaps, hand creams, and disinfectants. Though mainly considered an abiotic inhabitant, P. aeruginosa was also found as part of the human flora of gut, arms, perineum, throat, and nose, and can also become an opportunistic pathogen (Moore and Flaws 2011; Pachori et al. 2019). Also, the lung of most of cystic fibrosis (CF) patients are typically infected with P. aeruginosa, accompanied by poor response to antimicrobial therapy (Lyczak et al. 2002).
Strains belonging to P. aeruginosa are at the core of healthcare-associated infections (HAI), leading to endemics of high morbidity. It is reported that 10% of all HAI are attributed to P. aeruginosa, and mainly affect immunosuppressed and intensive care unit patients, with severe cases of pneumonia and bacteremia (Raman et al. 2018; Pachori et al. 2019). Wistfully, 32.1% of P. aeruginosa isolates in Europe were reported resistant to at least one of the regular antimicrobial groups (ECDC 2019). Indeed, a group of the so-called “high-risk clones” of multidrug-resistant P. aeruginosa poses an extreme threat to be treated with urgency, as no available therapies are at hand. Also, previous antibiotic therapies are now blamed for the emergence of carbapenem-resistant P. aeruginosa. As a result, the implementation of non-antibiotic treatments and of mixed therapies to minimize the employed titers of antibiotics, are considered is imperative (Raman et al. 2018).
Contaminated tap water, sinks, plumbing systems, etc. are familiar sources for the resistant microbial spread of HAI from P. aeruginosa, usually due to cross-contamination from wastewater and poor hygiene protocols. In these spots, stagnant water is blamed for being a natural incubator for P. aeruginosa biofilm growth (Breathnach et al. 2012; Quick et al. 2014; Bicking Kinsey et al. 2017). Biofilm is a microbial community embedded in an extracellular polymeric substance (EPS) and is considered the dominant mode of microbial life (Stoodley et al. 2002). EPS is mainly composed of polysaccharides, proteins, lipids, and nucleic acids, providing the microbes with a well-hydrated environment for three-dimensional growth. Biofilms protect bacteria from ultraviolet radiation, extreme temperature, pH, salinity, and antibacterial factors. Besides the presence of EPS as a physical barrier, biofilm physiology can contribute to gaining resistance to multiple antibiotics (Yin et al. 2019). Among the several molecular components that are crucial for bacterial adaptability and biofilm formation, functional amyloids—a group of loosely ordered peptides—are recognized in both bacteria and fungi to be implicated in virulence and biofilm formation (Van Gerven et al. 2018). Amyloids are characterized by a signature motif of usually extracellular, elongated, unbranched fibers consisting of stacked β-sheets. These formations are highly resistant to protease treatment and are routinely identified by their ability to bind the dyes thioflavin T (ThT) and Congo red (CR) (Eisenberg and Jucker 2012; Taglialegna et al. 2016). Active metabolism is recognized as imperative for functional amyloid regulation and biofilm establishment (Reshamwala and Noronha 2011).
The functional amyloid of P. aeruginosa (Fap) has been recognized by its ability to bind both ThT and CR (Larsen et al. 2007; Dueholm et al. 2010). Fap fibers (amyloid-like fimbriae, ALF) are coded by the fap operon (fapABCDEF), which is responsible for the expression of six different constituents of ALF. Fap subunits are chaperoned and processed at the extracellular space by the activity of FapA, D, and F, where polymerization is performed on the surface of the bacterial cell wall. FapC is recognized as the main component of these fibers, while FapB acts as an amyloid nucleator for FapC polymerization. ALF are considered essential for the biofilm formation of P. aeruginosa, and fap expression leads to bacterial aggregation and increased biofilm formation (Dueholm et al. 2013; Rouse et al. 2016; Rasmussen et al. 2023). ALF are highly resistant to high temperatures, dryness, and detergents and thus provide bacteria with the proper environment for survival in impossible environmental and in-host circumstances (Dueholm et al. 2010; Zeng et al. 2015).
Serrapeptase (SPT) is a metalloprotease discovered in the Gram-negative bacterium Serratia marcescens (Miyata et al. 1970). Multiple beneficial properties have been attributed to SPT, namely anti-inflammatory, analgesic (Tachibana et al. 1984; Nakamura et al. 2003; Al-Khateeb and Nusair 2008), anti-amyloid (Fadl et al. 2013; Metkar et al. 2017, 2020) and anti-biofilm properties (Longhi et al. 2008; Artini et al. 2011; Papa et al. 2013; Selan et al. 2015, 2017; Zapotoczna et al. 2015; Tsitsa et al. 2016). SPT is a promising agent for pharmaceutical applications, as it is non-toxic for animal cells (Chopra et al. 2009; Papa et al. 2013; Selan et al. 2017). Also, SPT may boost antibiotic efficacy in humans (Passariello et al. 2012; Sannino et al. 2013) and rats (Mecikoglu et al. 2006), presenting no side effects during its employment, except for some gastrointestinal disturbances. Though reduced bioavailability was highlighted as a key drawback for the medical employment of SPT (Jadhav et al. 2020), SPT-loaded nanoparticles, liposomes, gels, etc. can be employed for optimizing SPT's in vivo administration (KV et al. 2008; Shinde and Kanojiya 2014; Devlin et al. 2021). However, the possible anti-biofilm effect of SPT on P. aeruginosa and its mechanistic details has been scarcely studied (Artini et al. 2022).
We have previously proven that SPT can be exploited against biofilm formation from both susceptible and multi-drug resistant Staphylococcus aureus strains; furthermore, we have demonstrated the effect of SPT against the bacterial viability, amyloids, and virulence factors (Katsipis and Pantazaki 2023). Here, the anti-biofilm activity of SPT against P. aeruginosa ATCC 27853, a bloodborne model-strain with multiple antibiotic resistances (Cao et al. 2017), was verified in a semi-quantitative manner. In the present study, we also attempted to evaluate the biofilm formation from P. aeruginosa and the effect of SPT on it with diverse methods, e.g., plastic tissue culture plate (TCP) method and optical, fluorescence, and confocal microscopy. As far as we know, the anti-biofilm activity of SPT against this bacterial strain has not been reported before. Furthermore, the inhibitory effect of SPT on bacterial viability/metabolic activity is also demonstrated. The effect of SPT on functional amyloids in terms of ThT- and CR-positive compounds has been also studied, while in silico molecular docking calculations on the interaction of SPT with crucial peptides FapC and FapD provide probable further insights on the anti-amyloid potential of SPT. Statistical analyses have also been used to study the interrelation of Pseudomonas viability and amyloid titers with biofilm formation. The present results should offer novel possibilities for introducing SPT in clinical practice and demonstrate a mechanistic model for its activity against biofilm and amyloids of P. aeruginosa.
Materials and methods
Reagents and microbial strains
SPT was purchased as capsules of 60,000 IU (Health Aid LTD, UK). The powdered interior of the capsules was mixed with growth medium (1 capsule interior per 10 mL medium), vortexed, and filtered (Minisart NY 25, sterile 0.45-µM filters, Sartorius Stedim Biotech GmbH, Goettingen, Germany) to abort non-dissolved materials and excipients. No further purification process was employed. The SPT concentration of the received solutions was estimated with the Bradford-Bearden assay, as modified by Zor and Selinger (Bearden 1978; Zor and Selinger 1996). At the same time, the proteolytic efficiency of the preparations was tested with the protease substrate azocasein (A-2765, Sigma-Aldrich, St. Louis, IL, USA), as previously described (Vélez-Gómez et al. 2019). Tryptone (#403682), dimethyl-sulfoxide (DMSO) (#D5879), and (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) (MTT) (#A2231) were purchased from PanReac AppliChem (Darmstadt, Germany). Soybean peptone (#70,178), crystal violet (CV) (#C0775), ThT (#T3516), 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (#D9542), and CR (#75768) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sterile double-distilled H2O was used throughout the experimental procedures. P. aeruginosa strain ATCC® 27853™ (Boston 41501) was used in this study.
Growth conditions and biofilm formation
All growth media were autoclaved before use. Bacteria were stocked at 20°C in Luria–Bertani (LB) medium (% w/v: 1 tryptone, 0.5 NaCl, 0.5 yeast extract) containing 20% (v/v) filtered glycerol. Initial cultures were received with the suspension of 100 µL of stock culture in 10 mL of LB medium and grown overnight, at 37°C, in a shaking incubator. The composition of the employed medium for biofilm growth was defined after pilot tests with several dilutions of LB or Tryptic Soy Broth (TSB) medium for the determination of planktonic growth, biofilm formation, and functional amyloids (see the following paragraphs of the “Materials and methods” section for the determination protocols). Biofilm formation was finally performed with a 4 times diluted TSB (% w/v: 0.425 tryptone, 0.075 soybean peptone, 0.125 NaCl, 0.0625 Κ2HPO4), supplemented with 0.25% (w/v) glucose, for 24 h, at 37 °C, at static conditions.
For the semi-quantification of biofilm formation, bacteria were grown in polystyrene (PS) plastic surface of 96-well TCP, in the presence (Treated) or absence (Control) of SPT, while for microscopy evaluation bacteria were grown on glass microscopy slides (1 × 1 cm). Overnight LB cultures were diluted 20 times with biofilm medium and transferred in 96-well TCPs (#CLS3997, Corning, New York. USA) for TCP method, or petri dishes for microscopy evaluation, respectively. Growth on glass slides was performed by immersing the slides in the diluted bacteria in the petri dishes. For planktonic bacteria estimation, the cultures’ turbidity was read at 630 nm in a microplate reader (BioTek Instruments Inc., USA). Biofilm determination on the surface of the TCP wells was performed after dyeing the cells with CV, as previously described (Katsipis et al. 2021). Microscopy evaluation of CV-stained glass slides was done after fixing the biofilms with 2.5% (v/v) glutaraldehyde for 15 min, followed by staining with 0.4% (w/v) CV for 10 min. Stained biofilms were then photographed under the bright field of a Nikon Eclipse Ci fluorescence microscope.
Biofilm evaluation by confocal microscopy
Analysis of biofilm-coated surfaces was performed according to standard ISO 25178 using a 3D Optical Surface Metrology System Leica DCM8 device. The biofilm-coated surfaces, once removed from the incubation test cells, were washed off with PBS solution. Afterward, glutaraldehyde solution (2.5%, v/v) was applied to the surface biofilm fixation as mentioned above, enabling the analysis of its morphology. For each biofilm-coated surface, three images were obtained (× 10 magnification), corresponding to a surface of 1.3 mm × 1.6 mm for each image. Images were subjected to analysis using the Leica Map software (LeicaGmbH, Germany), and the parameters of the roughness profile (Sdr, %), the maximum height of peaks (Sp, µm), and the arithmetic mean height of surface (Sq, µm) were determined using the acquired data.
Determination of bacterial viability by MTT assay
MTT assay is now accepted as an effective method for estimating bacteria viability and metabolic activity (Wang et al. 2010; Grela et al. 2018). P. aeruginosa bacteria were grown in the presence or absence of SPT, in glass tubes sealed with a gauze cap filled with hydrophobic cotton, and with the biofilm conditions described before. After growth, bacteria were harvested by centrifugation (5,000 rpm for 10 min) and then washed and resuspended in PBS. The viability of the bacteria cells was determined with 0.5 mg/mL ΜΤΤ (final concentration), after incubation at 37°C, for 30 min, under mild shaking. A sample containing PBS and the MTT substrate was also prepared as blank. The formazan crystals received due to MTT reduction were then centrifuged and dissolved with DMSO, and the absorbance was read at 570 nm in a microplate reader. The bacterial viability/metabolic activity was finally normalized against the total bacterial turbidity of the PBS-resuspended cell samples and was expressed by setting the viability of the untreated bacteria at 100%, as previously described (Wang et al. 2010).
Estimation of functional amyloid content with Thioflavin T
In order to study the distribution of functional amyloids on the biofilm surface, as well as the effect of SPT treatment on them, P. aeruginosa was grown on glass slides and then fixed with glutaraldehyde. Afterward, slides were covered with a PBS solution containing 10 µΜ ThT for amyloid staining and 1 µg/mL DAPI for general EPS staining due to extracellular DNA complexation. Staining was performed for 15 min in the dark, and then biofilms were washed thoroughly with PBS and observed under a Nikon Eclipse Ci fluorescence microscope equipped with a Color Camera Nikon DS-Fi3 (× 20, FITC filter, 80 ms exposure for ThT/ × 20, DAPI filter, 30 ms exposure for DAPI).
Image processing
Image analysis was carried out employing the open-access software package Fiji (Image J, US National Institutes of Health, Bethesda, Maryland, USA). Briefly, regarding the processing, the experimental images were converted to greyscale, and subsequently, a threshold was set to create binary images separating the background from features of interest. Finally, analysis of particles was performed providing information about the total surface coverage (%) and their number. More specifically, concerning the fluorescence imaging processing, brightness, and contrast for green and blue channels, respectively, were also adjusted using Fiji, while both channels were merged to generate composites. For each biofilm-coated surface, at least 6 images corresponding to different points of the surface were obtained and further analyzed statistically.
Estimation of functional amyloid content with Congo red
P. aeruginosa amyloids were estimated with the CR assay, as previously described (Reichhardt et al. 2015). Briefly, bacteria grown in glass tubes under biofilm conditions as described above were received and mixed with a chill CR solution of 10 µg/mL (final concentration), in PBS. A sample in the absence of bacteria was also prepared to estimate the absorbance of the pure dye solution. Bacteria were incubated at room temperature for 10 min and then pelleted by centrifugation (13,500 rpm for 5 min). The absorbance of the supernatant for CR was then read at 500 nm with a Selecta 2005 UV–Vis spectrophotometer (cell path of 1 cm). The retained CR dye from bacteria corresponding to the amyloid compounds was inversely calculated, by subtracting the absorbance values from the ones received from samples containing only the dye solution. Finally, results were normalized versus the rough bacterial turbidity of the PBS-resuspended cell samples and were expressed by setting the amyloid content of the untreated bacteria at 100%.
Molecular docking calculations
To elucidate the possible interaction between SPT and the FapD or FapC proteins, the pyDockWEB server (https://life.bsc.es/pid/pydockweb) was employed (Jiménez-García et al. 2013) for structural prediction of protein–protein interactions. Given the 3D coordinates of two interacting proteins, pyDockWEB returns the best rigid-body docking orientations generated by FTDock and evaluated by the pyDock scoring function, which includes electrostatics, desolvation energy, and limited van der Waals contribution. PyDockWEB server makes use of the SCWRL software for rebuilding missing side chains. PyDockRST software package uses the percentage of satisfied distance restraints, together with the electrostatics and desolvation binding energy, to identify correct docking orientations. This methodology drastically improves the docking results when compared to the use of energy criteria alone and is able to find the correct orientation within the top 20 docking solutions in 80% of the cases (Chelliah et al. 2006). From the output sorted by the pyDock scoring function, the top one hundred conformations was derived among a total of 500 conformations. From a table with a list of generated conformations scored and ranked by the energy scoring, the best protein complexes between SPT and FapC or, FapD were selected.
For FapC Pseudomonas sp. UK4 FapC amyloid-like fimbriae precursor protein was used, with accession coding EEP64551.1 of European Nucleotide Archive (ENA) Advanced Search API (https://www.ebi.ac.uk/ena/browser/home) in FASTA format for modeling and prediction of the protein by running the BLAST sequence similarity search by UniProt (https://www.uniprot.org/uniprotkb/C4IN70/entry) with very good Model Confidence (most of the structure displaying pLDDT values among 70 and 90 (Confident), while lower portions of the structure display either pLDDT > 90 (Very high) or 70 > pLDDT > 50 (Low) and pLDDT < 50 (Very low)) (Protein name: FapC amyloid-like fimbriae protein; Gene name: fapC; ORF name: PSUK4_00030; Organism name: Pseudomonas sp. UK4; Taxonomic identifier: 452,680 NCBI; Length 250; Mass (Da) 24,998). The AlphaFold structure prediction database (either from the AlphaFoldDB databases or the UniProt databases) was downloaded for the FapC 3D structure model AF-C4IN70-F1-model_v4.pdb (https://alphafold.ebi.ac.uk/entry/C4IN70).
On the other hand for FapD, the modeled after the homologous C39 peptidase domain of ABC transporter PCAT1 was employed, PDB: 4RY2 (FapD model of conserved core domain) (Dueholm et al. 2010).
Statistical analyses
All statistical analyses and graph constructions were done with GraphPad Prism 8 (GraphPad Software Inc.). Provided graphs include bars representing mean values ± standard error of the mean (SEM), from at least three independent experiments. The possible statistical significance for differences between the untreated (control) and treated samples using increasing SPT concentrations, was examined with standard one-way analysis of variance (ANOVA), after correcting for multiple comparisons with Dunnett’s test. Before performing ANOVA, the normality of the replicates was verified with Anderson–Darling, D'Agostino-Pearson omnibus, Shapiro–Wilk, and Kolmogorov–Smirnov tests, while possible significant differences between the standard deviations of the various samples were evaluated with Brown-Forsythe test. Inhibitions at 50% of the untreated sample (IC50) and the corresponding 95% of confidence intervals – CI (95%) were calculated based on the equation: log [SPT] vs. normalized inhibition (%), with variable slope. Possible correlations were examined with Pearson’s analysis, determining r coefficients and two-tailed p values. Statistically significant results were considered for p < 0.05. Notations for statistically significant differences between control (untreated) and treated samples: *p < 0.05; **p < 0.01;*** p < 0.001; and ****p < 0.0001.
Results
The effect of growth medium composition on biofilm formation of P. aeruginosa
The composition of the growth medium was standardized in terms of efficient biofilm growth. In the current growth conditions and the LB medium, P. aeruginosa has mainly adapted to the planktonic growth mode, as proven by the very low CV staining. To that notice, the LB medium was not studied further. TSB medium supplemented with 1% (w/v) glucose was evaluated for biofilm formation. Additionally, two different TSB dilutions were also analyzed: 2 times and 4 times diluted. The planktonic bacteria growth and the levels of amyloids were also analyzed for each medium dilution employed. The results for TSB are graphically depicted in Supplementary Fig. S1.
Increased biofilm formation was found at the highest studied dilution of the growth medium (4 times diluted TSB), implying that in the studied conditions, lower availability of nutrients induces a biofilm-growth phenotype for P. aeruginosa in a greater degree compared to a nutrient-rich environment. Increased levels of biofilm formation were accompanied by lower levels of planktonic bacteria growth, as demonstrated by the turbidity of the growth medium, and elevated titers of functional amyloids, as determined by CR staining. These results indicate the induction of functional amyloid expression and biofilm formation under lower availability of nutrients. Consequently, 4 times diluted TSB was employed for all further experimentations.
SPT inhibits the biofilm formation of P. aeruginosa on plastic
The ability of various SPT concentrations to inhibit the formation of P. aeruginosa biofilm on TCP wells was studied under static conditions. Bacterial biofilms were stained with CV—a dye that binds mainly on polysaccharides like peptidoglycans (Jones 2017) and semi-quantified after the extraction of the dye and its determination at 570 nm. After normalization of the data, the ability for biofilm formation (% of the control sample) on plastic surfaces of TCP is presented in Fig. 1a for indicative SPT concentrations of 0.25 to 10 µg/mL. The analysis with the TCP method demonstrated that SPT is a potent inhibitor for biofilm formation of P. aeruginosa on plastic surfaces, presenting a maximum inhibition at 10 µg/mL of SPT (-55%) and a calculated IC50 value of 11.26 µg/mL (CI 95%: 7.95 to 20.14 µg/mL).
Inhibition of biofilm formation of Pseudomonas aeruginosa by serrapeptase (SPT) and effect on planktonic bacteria. P. aeruginosa ATCC 27853 bacteria were grown under static conditions in tissue culture plates (TCPs) in the presence or absence of SPT. Biofilm formation (a) was semi-quantified with crystal violet staining after dye extraction and read at 570 nm, while the levels of planktonic bacteria (b) were estimated by determination of medium turbidity at 630 nm. Biofilm inhibition correlates inversely with the rough density of planktonic bacteria (c). P. aeruginosa ATCC 27853 bacteria grown under static conditions in glass tubes, in the presence or absence of SPT, were collected and analyzed for their ability to reduce (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) (MTT). The levels are presented as % of the control (untreated) value (d). Biofilm inhibition correlates with reduction in planktonic bacteria viability (e). Bars represent mean values ± SEM from at least three independent experiments, with the value of the untreated bacteria culture (control) set at 100%. Standard ANOVA with Dunnett’s correction for multiple comparisons was employed for the statistical analysis. Notations for statistically significant differences between control (untreated) and treated samples: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
To further demonstrate the anti-biofilm efficacy of SPT, the bulk of planktonic bacteria were estimated at the growth medium by measuring its turbidity at 630 nm. The levels (% of the control sample) of planktonic bacteria for each SPT treatment and the control are presented in Fig. 1b. Increasing doses of SPT lead to higher floating bacteria titers. However, statistical significance could not be verified. However and in agreement with biofilm diminishment under SPT treatment, a negative, significant correlation between biofilm formation and planktonic bacteria turbidity was verified (Fig. 1c), indicating that SPT treatment possibly prevented the ability for growth under biofilm conditions and led to higher numbers of planktonic bacteria. However, it is not possible to determine by this turbidity measurement if the floating bacteria were viable and thus, a viability assay was also employed.
SPT treatment impairs the viability/metabolic activity of P. aeruginosa
MTT is reduced mainly by oxidoreductive enzymes of the oxidative metabolism of viable bacteria (Grela et al. 2018) and thus can be exploited for studying the effect of SPT on P. aeruginosa viability and metabolic capability. Bacteria were grown under biofilm conditions, collected, and then examined for their ability to actively reduce MTT. After normalization, test results are summarized in Fig. 1d.
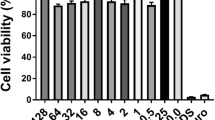
SPT treatment causes a significant decline in the metabolic efficiency of P. aeruginosa bacteria in a dose-dependent manner, with a calculated IC50 value of 3.07 µg/mL (CI 95%: 2.26 to 4.13 µg/mL). The maximum viability inhibition was -63% when 10 µg/mL of SPT was employed. Correlation analysis also indicated that impairment of metabolic viability coincides with biofilm inhibition, as a significant positive correlation was found (Fig. 1e). These results demonstrate that the presence of SPT can hijack proper bacterial metabolism, leading to impaired viability. This effect can and possibly sensitizes bacteria and thus presents a promising potential for antimicrobial therapy.
SPT inhibits the biofilm formation of P. aeruginosa on glass
The ability of SPT to inhibit the formation of P aeruginosa biofilm on glass was also studied. Bacterial biofilms grown on microscopy slides were stained with CV and then observed and photographed under a bright field. Representative images received from the analysis are provided in Fig. 2a. Untreated P. aeruginosa cultures form a dense network consisting of bacterial aggregates and intermediate abiotic areas, that probably represent water channels for biofilm active nourishment. However, treatment with 2, 5, or 10 µg/mL of SPT dramatically disrupted biofilm structure, with only minimal bacterial assemblies observed at the highest employed dose. Densitometric analysis on the received images from CV-stained biofilms further supports the anti-biofilm ability of SPT, proving that protease treatment can abruptly impair biofilm colonization on the glass surface (IC50 = 0.27 µg/mL, CI 95%: 0.06 to 0.55 µg/mL) (Fig. 2b). A maximal inhibition of -92% was recorded at 10 µg/mL of SPT treatment. Additionally, SPT led to a significant dose-dependent decrease of the microbial aggregates, in terms of calculated particles on the observed fields (Fig. 2c). It should be noted that due to the dense biofilm network of untreated biofilms, it was technically problematic to estimate the rough number of microbial aggregates with the particle analysis, so the results from untreated samples were omitted from the given graph and statistical analyses were performed versus the lower employed dose of 2 µg/mL SPT. These results prove the preventive nature of SPT against biofilm formation on glass surfaces, which, based on the current results of the TCP method, surpasses the corresponding effect for plastic surfaces.
Pseudomonas aeruginosa biofilms formed on glass slides in the absence (untreated) or presence of SPT concentrations (2, 5, and 10 µg/mL), after fixation and staining with crystal violet. Bright images (a) are received from Nikon Eclipse Ci fluorescence microscope. Densitometric analysis of the coverage of glass surface (b) and number of particles for different SPT concentrations (c) were estimated from analysis of the received images with Image J. Due to the dense biofilm network of untreated biofilms, particle analysis from untreated samples were omitted from the given graph and statistical analyses were performed versus the lower employed dose of 2 µg/mL SPT. Bars represent mean values ± SEM from at least three independent experiments. Standard ANOVA with Dunnett’s correction for multiple comparisons was employed for the statistical analysis. Notations for statistically significant differences: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Confocal microscopy of fixed Pseudomonas biofilms on glass slides provided further understanding of the effect of SPT and representative analyses graphs are provided in Fig. 3. The addition of SPT induced significant modifications in the morphology of biofilms on glass. 3D images suggested a much thicker and denser biofilm matrix with numerous peaks higher than 3 µm and various scattered peaks greater than 5 µm. SPT treatment provoked the disruption of biofilm formation and structure formulation. As shown in Fig. 3, biofilm thickness was considerably lowered after the application of 2 µg/mL SPT, while higher doses up to 10 µg/mL caused the formation of irregular structures with sporadic peaks that did not exceed 2.5 µm. Profilometric measurements over the majority of glass surfaces confirmed the reduction of surface roughness, in terms of developed surface area. Indeed, SPT increases up to 5 µg/mL minimize surface developed area close to clean surface. Likewise, maximum height of peaks and mean height of the surface were drastically reduced even after treatment with just 2 µg/mL SPT (Supplementary Figures S2 and S3).
SPT treatment reduces functional amyloid levels of P. aeruginosa
The dyes CR and ThT were previously reported for employement in the determination of bacterial functional amyloids (Zeng et al. 2015; Reichhardt et al. 2015). Given the significance of Fap for aggregation and biofilm formation of Pseudomonas (Dueholm et al. 2013) and the anti-amyloid properties attributed to SPT (Metkar et al. 2017), the effect of SPT on P. aeruginosa amyloids grown under biofilm conditions was studied here. Cells were stained with the fluorescent dye ThT for amyloid detection and with DAPI as a general marker for biofilm EPS and observed under fluorescence microscope (Fig. 4a) or stained with CR and the retained dye was inversely estimated. The normalized results for the titers of ThT- and CR-positive compounds are presented in Fig. 4b and c, respectively.
Effect of serrapeptase (SPT) on levels of functional amyloids of P. aeruginosa bacteria. (a) P. aeruginosa ATCC 27853 bacteria were grown under static conditions on glass coverslips, in the absence (untreated) or presence of different SPT concentrations, were fixed with glutaraldehyde, and stained with thioflavin T (ThT) for functional amyloids and 4′,6-diamidino-2-phenylindole (DAPI) for rough staining of biofilm matrix due to extracellular DNA. Bacteria were then observed under a Nikon Eclipse Ci fluorescence microscope (× 20, FITC filter, 80 ms exposure for ThT, × 20, DAPI filter, 30 ms exposure for DAPI) and the density of the stained cells was quantified with Image J (b). P. aeruginosa ATCC 27853 bacteria were grown under static conditions in tubes in the presence or absence of SPT and were collected and mixed with CR solution. The retained dye, which corresponds to the levels of expressed functional amyloids, was inversely estimated by reading the absorbance of the supernatant at 500 nm, after discarding bacteria with centrifugation. The levels are presented as % of the control (untreated) value (c). Bars represent mean values ± SEM from at least three independent experiments, with the value of the untreated bacteria culture (control) set at 100%. Standard ANOVA with Dunnett’s correction for multiple comparisons was employed for the statistical analysis. Notations for statistically significant differences between control (untreated) and treated samples: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Pearson’s correlation analysis (inset of c) indicated a significant, positive correlation between biofilm formation and bacterial viability
Treatment with SPT leads to a dose-dependent reduction of functional amyloids of P. aeruginosa bacteria, with an IC50 value estimated at 10.07 µg/mL (CI 95%: 7.77 to 14.68 µg/mL) for CR-positive compounds, while ThT-positive compounds were also found to be reduced (in terms of ThT/DAPI ratio), with a similar IC50 of 8.64 µg/mL (CI 95%: 6.65 to 11.33 µg/mL). Maximum inhibition of amyloid levels was recorded at 10 µg/mL of SPT, at -48% to that of control for CR-positive compounds and at -52% for ThT-positive compounds. Correlation analysis (inset of Fig. 4c) indicated that amyloid reduction, as determined by CR retention, interrelates positively and significantly with biofilm inhibition. These results verify the importance of amyloids for adequate biofilm formation of P. aeruginosa, as a lower amyloid content in P. aeruginosa is directly linked with a lower capability for biofilm formation. Moreover, they underline the anti-amyloid activity of SPT and propose a possible mechanism for that.
Protein–protein molecular docking calculations
Since SPT acts against bacterial biofilms with a possible effect on amyloids of P. aeruginosa and, both FapC and FapD are supposed to be possible targets of SPT, it is most rational to seek any existing interaction between SPT and any of these two peptides to suggest a probable proteolytic mechanism. The total binding energies of the best binding pose predicted by pyDockWEB (conformation of first rank according to the computed binding energy) between SPT and FapC and FapD proteins (Table 1), were calculated taking into account the following energetic components: electrostatic, desolvation, and Van der Waals (VDW) (term weighted to 0.1 by default). The total binding energy represents the sum of the 3 previous energies. The proposed structural prediction models of two Fap fibril biogenesis, the major Fap amyloid component FapC amyloid–like fimbriae protein produced by the BLAST sequence similarity search by UniProt model of FapC (AF-C4IN70-F1-model_v4.pdb) and the FapD modeled after the homologous C39 peptidase domain of ABC transporter PCAT1 (PDB ID: 4RY2) are depicted in Supplementary Figure S4. For FapC the generated model demonstrated very good Model Confidence (most of the structure displaying pLDDT values between 70 and 90 (Confident), while lower portions of the structure display either pLDDT > 90 (Very high) or 70 > pLDDT > 50 (Low) and pLDDT < 50 (Very low)).
The binding architecture models of protein–protein binding poses between SPT and FapC and between SPT and FapD are illustrated in Figs. 5 and 6. The docking predicts the formation of a variety of interactions between the components of the protein complexes including hydrogen bonds, hydrophobic, polar, π-polar, π-π displaced and T-shaped, π-alkyl hydrophobic, and π-anion charged electrostatic interactions (Supplementary Table S1 and Supplementary Table S2).
(a) A mechanistic model of the interaction between SPT and FapC demonstrated a possible binding/proteolytic activity of SPT on FapC. Binding architecture of FapC/SPT protein complex in the extracellular space at Binding Site I, after transportation of FapC (the main fibril component) out of the cell through the outer membrane using FapF trimeric β–barrel polypeptide membrane transporter via FapF pore. Proteins are depicted as cartoon colored by raspberry and light pink in the periplasmic and extracellular space, respectively (FapC), yellow–orange (SPT), and light pink, deep purple, and deep salmon (FapF for chains A, B, and C, respectively). FapC in the periplasmic space and FapF are additionally depicted with semi–transparent surface colored also by the cartoon colors. A close–up view of the Binding Site I mapping interactions of the best docking pose orientation, between SPT and FapC is also shown at the upper part of the figure, with the proteins depicted in cartoon representation and binding residues of both proteins illustrated in stick model colored according to cartoon. Binding contacts between SPT/FapC are indicated in yellow dotted lines. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics System. (b) A mechanistic model of the interaction between SPT and FapC at Binding Site II, demonstrating a possible binding/proteolytic activity of SPT on FapC. Binding architecture of FapC/SPT protein complex in the extracellular space at Binding Site II: the catalytic active site cleft ligated by Hisl76, Hisl80, Hisl86, and Tyr216. Proteins are depicted as cartoon colored by split pea green (SPT) and dirty violet (FapC) with additional depiction of semi–transparent surface colored also by the cartoon colors. The residues in the catalytic active site on the cartoon representation and the surface, are highlighted in firebrick color. A close–up view of the Binding Site II mapping interactions of docking pose orientation (at the upper right part of the figure), between SPT and FapC is also shown with the proteins depicted in semi–transparent surface representation and the binding residues of both proteins illustrated in stick model colored according to surface. Binding contacts between SPT/FapC are indicated in yellow dotted lines for SPT and purple for FapC. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics System
A mechanistic model of the biogenesis machinery of Fap in P. aeruginosa depicting the role of FapD in affecting the release of FapC to the extracellular space and the role of SPT binding to FapD on the FapC release. FapC passes through the outer membrane using FapF trimeric β–barrel polypeptide transporter (PDB: 5O67) via FapF pore. FapD, modeled after the homologous C39 peptidase domain of ABC transporter PCAT1 (PDB ID: 4RY2), is a protease involved in the proteolysis of Fap proteins and is shown to perform an essential proteolytic modification of FapC while residing in the periplasmic space between the inner and outer membrane, before its secretion from the cell. This activity may be affected by the SPT binding on FapD’s catalytic center, probably affecting its active peptidase function (the inhibitory activity of SPT on FapC release is shown by the inhibition arrow in the figure). The catalytic center of FapD is illustrated in the interface region between FapD and SPT in yellow–orange color. All proteins are depicted in cartoon colored in deep blue and violet purple for SPT and FapD, respectively, and in tv blue, purple blue, and deep purple for FapF (for chains A, B, and C, respectively). A close–up view of the binding site mapping interactions of the best docking pose orientation between SPT and FapD is also shown at the bottom right side of the figure, with binding residues of both proteins illustrated in stick model colored according to cartoon. Binding contacts between SPT/FapD are indicated in yellow dotted lines. The final structure was ray–traced and illustrated with the aid of PyMol Molecular Graphics System
The Predicted Aligned Error (PAE) plot for FapC amyloid-like fimbriae protein indicating the AlphaFold’s expected distance position error at residue x, when the predicted and true structures are aligned on residue y, is depicted in Supplementary Figure S5. From Supplementary Table S1 it is deduced that most binding contact residues of FapC correspond to regions involving residues 12–19 and 183–226. From Figure S5, the alignment of these residues x (12–19, 183–226 when the prediction and true structure are aligned on residue y) results in confidence in the relative position of them. From the PAE plot, it is deduced that the predicted errors for FapC are restricted to very low levels (dark green shades indicating low errors).
The docking procedure revealed two binding sites of the docked proteins SPT and FapC, namely Binding Site I (Fig. 5a) and Binding Site II (Fig. 5b) with similar total binding energies. The binding of FapC on SPT in Binding Site I is mediated in two distinct binding regions, the first in the non-repeat helical segment, and the second in the three imperfect R3 repeat sequences of FapC stacked on top of each other, along with the additional β-strands linker region (only the β-sheets connecting the R3 repeat sequences) formed inside the amyloid core (Fig. 5a). The first binding region of FapC on site I is revealed to be in contact with the C–terminal region of SPT and especially with α–helix G (the only helix in the C–terminal region) and the loops connecting the three–stranded antiparallel β–sheets β20, β21, and β22. As for the second region, it is found in contact with part of the N–terminal proteolytic domain of SPT and especially with α–helix B and the coils linking helices A and B and helix B with β–sheet β1. All binding contacts between SPT and FapC are mediated by residues belonging to the R3 repeat sequence of FapC and the helical region. The binding of FapC on SPT in Binding Site II includes the catalytic active site cleft ligated by three histidine residues Hl76, Hl80, and Hl86, and one tyrosine, Y216, completing thus the catalytic tetrad (Supplementary Table S1 and Fig. 5b).
The binding interface region between FapD and SPT is in the active catalytic center of FapD, indicating a possible proteolytic interference in the active peptidase function of FapD, beyond its processing chaperone activity (Fig. 6). Binding of SPT to FapD affects the proteolytic modification of FapC exerted by FapD while residing in the periplasmic space between the inner and outer membrane, before its secretion from the cell.
Discussion
The emergence of the so-called “superbugs” is potentially the “Phantom Menace” of future medicine. The extensive use of antibiotics and antibacterial agents in the past has raised a whole generation of multi-resistant bacterial strains, which will probably dominate the interest of many future generations of researchers and clinical practicians. These strains will allegedly cause a plethora of impossible-to-treat infection cases, especially inside the nosocomial environment, that will make the public expenses for healthcare and mortality rates explode to unprecedented heights (Dadgostar 2019). Consequently, it is of imperative need to implicate novel medications against microbial agents other than antibiotics, in order to minimize the employed doses, if not avoid their use at all (Bush et al. 2011).
One of the mechanisms for the multi-gaining of antibiotic resistance is tragically the most frequent mode of microbial growth: biofilms. Biofilm-growing bacteria cause chronic infections, characterized by persistent inflammation and tissue damage, while actively obstructing antimicrobial compounds and the host immune system. Notably, biofilm-growing bacteria may be up to 100- to 1000-fold more resistant to antibiotics, in comparison with planktonic bacteria (Høiby et al. 2010). The EPS of biofilms is actively contributing to resistance acquisition, by neutralizing or diluting antimicrobial substances. At the same time, biofilms can thrive through slow growth and the building of defences that create more and more resistant clones. Nutrient depletion and reduced oxygen levels may also contribute to the matter (Percival et al. 2015).
In the current study, P. aeruginosa biofilms have been studied. P. aeruginosa is one of the core species at HAI, characterized naturally by numerous resistances (ECDC 2019). Also, P. aeruginosa is the main pathogen for CF patients, who suffer from chronic infections with poor prognosis (Lyczak et al. 2002). P. aeruginosa in CF lungs forms biofilms with bacteria that are characterized by very slow growth (Yang et al. 2008). As antibiotics are usually active against doubling P. aeruginosa cells, their administration is generally failing to protect patients suffering from biofilm infections (Høiby et al. 2010). Thus, in the current study, the employment of an alternative anti-biofilm agent, namely SPT, has been attempted. SPT has been previously proven to be able to block or eradicate in vitro and/or in vivo biofilm formation from several bacteria, including Listeria monocytogenes (Longhi et al. 2008), Staphylococcus aureus (Artini et al. 2013; Papa et al. 2013; Selan et al. 2015, 2017; Devlin et al. 2021; Katsipis and Pantazaki 2023), and Staphylococcus epidermidis (Mecikoglu et al. 2006; Artini et al. 2013). The anti-biofilm effect of SPT on P. aeruginosa has not gained enough attention at present, with very few studies referencing its potential clinical employment (Selan et al. 1993; Passariello et al. 2012; Artini et al. 2022).
The anti-biofilm capacity of SPT against P. aeruginosa Boston 41501 (ATCC 27853) is demonstrated here—to the best of our knowledge—for the first time. Boston 41501 is a model strain isolated from a blood infection, presenting resistance against several antibiotic agents and disinfectants and employed commonly in studies of antibiotic resistance (Brown et al. 1995). In the current study, SPT was able to inhibit biofilm formation from the specific P. aeruginosa strain at both plastic and glass surfaces in a significant manner. Examination with confocal microscopy of the biofilms grown on glass has also revealed a much thicker and denser matrix, in comparison with biofilms grown in the presence of SPT.
In a recent study, Artini et al. have also confirmed the anti-biofilm efficacy of SPT against P. aeruginosa PA14 and several isolated clinical strains from CF lungs, impairing biofilm formation vaguely 40 to 70%. The authors proved that under the effect of SPT, P. aeruginosa presented impaired attachment to abiotic surfaces as well as reduced ability for adhesion/invasion of eukaryotic cells. Also, treatment with SPT decreased the virulence of the bacteria, as confirmed by altered pyocyanin and pyoverdine production, impaired swarming motility, and reduced staphylolytic protease production (Artini et al. 2022). However, the authors did not study the possible correlation of these parameters with biofilm formation and did not provide a possible mechanistic explanation for biofilm inhibition. Selan et al. worked on a combinational therapy with SPT and ofloxacin for several clinical strains of P. aeruginosa isolated from prosthetic device infections. The results proved that SPT could assist the microbiocidal effect of the employed antibiotic against both biofilm and planktonic bacteria (Selan et al. 1993). Several other studies also offered insights into the process, providing evidence that SPT can also enhance the activity of ofloxacin, azithromycin, ciprofloxacin, vancomycin, rifampicin and levofloxacin against S. aureus, S. epidermidis, and methicillin-resistant S. aureus (MRSA) (Selan et al. 1993; Thaller et al. 1997; Hogan et al. 2017; Gupta et al. 2017). Additionally, SPT was previously found to be able to be employed as a combined therapy of peri-implantitis, enhancing the success rates and providing better tissue repair around successfully treated tooth implants (Sannino et al. 2013).
Interestingly, our results have demonstrated that SPT can suppress biofilm build-up more strongly in glass slides (hydrophilic surface) than in plastic surfaces of TCP plates (hydrophobic surface), presenting an IC50 of 0.27 µg/mL and 11.26 µg/mL, respectively. Pereira et al. have verified that sessile growth of P. aeruginosa in plastic or glass surfaces induces differential biofilm phenotypes, with biofilms growing on plastic maturing at a faster pace than the ones in glass, with the latter presenting a more immature structure. The authors suggest that charge, hydrophobicity, and roughness of the surface would be important factors during biofilm development (Pereira et al. 2015). Additionally, in a multi-microbial biofilm study, P. aeruginosa showed comparatively higher hydrophobic surface properties than E. coli and S. aureus, which was regarded as significant for bacterial adhesion and directly proportional to biofilm formation (Mirani et al. 2018). Thus, it could be deduced that the highly hydrophobic area of TCP wells would strongly support steadier biofilm formations from P. aeruginosa, which could explain the need for significantly higher titers of SPT for obstructing its settling.
Low oxygen and nutritional requirements, slow growth, extended survival and antibiotic resistance gaining are recognized in P. aeruginosa. These conditions are mainly found in CF patient lungs, where bacteria thrive in a biofilm mode (Rossi et al. 2021). In the current study, the ability of P. aeruginosa to present increased levels of biofilm formation under nutritional deficiency has been verified. Previous reports demonstrate that glucose supports the growth of small, structured biofilms of P. aeruginosa than uniform biofilms, including less motile and swarming bacteria—a behavior attributed to quorum sensing control. In addition, P. aeruginosa grown with glucose express higher amounts of rhamnolipids—a surfactant that leads to biofilm dispersion and remodeling (Shrout et al. 2006), suggesting that in nutrient-rich environments, P. aeruginosa would preferably adopt a motile growth pattern.
To further study the effect of SPT, P. aeruginosa grown under the effect of SPT was examined for its metabolic viability. Results proved that SPT could ameliorate bacterial viability, in a way that coincides with biofilm inhibition, as also implied by correlation analysis. Recently, treatment of S. aureus biofilms with SPT has provided evidence for dysregulation of phosphate metabolism—a crucial factor for biofilm formation and bacterial viability (Danikowski and Cheng 2018; Katsipis et al. 2021; Katsipis and Pantazaki 2023). However, as far as we know, this is the first time that the effect of SPT on the viability of P. aeruginosa or any other Gram-negative bacteria is assessed with a standard method as the MTT test. Previously, Artini et al. have also proposed that impaired production of pyocyanin and pyoverdine due to SPT treatment is an effect correlating with reduced virulence—a factor the authors regard more important than viability inhibition (Artini et al. 2022). It should however be noted that the MTT assay used here is a rough measure of the bacterial metabolic activity, basically due to oxidoreductive enzymes (Grela et al. 2018). So, these results also provide evidence that SPT treatment may lead to impairments in bacterial homeostatic mechanisms that coordinate metabolism—an event forcing bacteria to abandon the biofilm environment. It is worth mentioning that SPT did not seem to impair the planktonic bacteria titers, as calculated by turbidity assays, and thus its possible toxic behavior seems not to be applied on motile than to sessile, biofilm-forming bacteria. However, it should be taken under notice that turbidity measurements cannot exclude dead bacteria. To that end, part or whole of the sum of the metabolically inactive bacteria may correspond to irreversibly non-viable cells. In a previous study of combinational therapy of several P. aeruginosa clinical strains with ofloxacin, SPT significantly reduced the minimal inhibitory concentration and minimal bactericidal concentration of the employed antibiotic, only regarding sessile bacteria, proving the thought that SPT specifically targets biofilm-forming bacteria (Selan et al. 1993). A following study assessing the ability of P. aeruginosa to grow new biofilms or colonies after SPT employment, would give more insight into the possible immediate and independent toxicity of this protease.
The functional amyloid titers, as estimated with CR retainment, seem to correlate with increased biofilm formation under low nutrition in the current study, as previously demonstrated for curli expression in several Enterobacteriaceae (Castelijn et al. 2012; Paytubi et al. 2017; Leech et al. 2020). Studies on the function of P. aeruginosa Fap and its expression are still in their first steps (Dueholm et al. 2010, 2013; Zeng et al. 2015). Fap expression is imperative for bacterial adaptation in CF lungs and biofilm formation and its overexpression is enough to cause a similar phenotype of lung infection by P. aeruginosa in mice non-carrying the CF-related allele (Beg et al. 2023). Also, Fap has been proven to increase the hydrophobicity and stiffness of Pseudomonas biofilms, providing better resistance against environmental stress, and providing better chances for successful colonization. Deletion of most of the Fap operon genes leads to the loss of these characteristics (Zeng et al. 2015). Also, FapC deletion – the main component for ALF buildup, impedes the virulence of P. aeruginosa, while deletion of other operon genes (except FapA), leads to lower aggregation and diminished biofilm formation (Dueholm et al. 2013).
Reduced amyloid titers in P. aeruginosa biofilms due to SPT treatment were verified in the current study, with both CR-retainment methodology and by observing and analyzing the ThT-binding of amyloids with fluorescence microscopy. As expected, attenuation of biofilm in the presence of SPT was firmly correlated with CR-staining, indicating that SPT could impair the early formation of ALF. The exact mechanism of this effect is yet to be discovered. It was previously demonstrated that SPT could impair the amyloid formation of insulin fibers—a model for anti-amyloid studies both in vitro and in vivo (Metkar et al. 2017, 2020). A proteolytic activity against polymerizing ALF could be expected, as SPT was previously reported to affect several extracellular peptides of S. aureus, in accordance with biofilm inhibition (Artini et al. 2011). However, a later report proved that the SPT effect on S. aureus biofilm could not be attributed to a proteolytic mechanism, as mutations affecting the active site of the enzyme did not impair its anti-biofilm efficacy (Selan et al. 2015).
In silico molecular docking studies also provided mechanistic insight into the understanding of the interaction between SPT and FapC or FapD, and demonstrated a possible binding/proteolytic activity of SPT on both peptides. During the fibril build-up, each FapC monomer is stabilized not only by interactions with other monomers in the fibril, but also by contacts between the individual imperfect repeats, which most likely constitute β–hairpin helices (Rouse et al. 2017). R3 repeats are indicated to play an important role in stabilizing the fibrils (Christensen et al. 2020). Interestingly, there is evidence that repeat sequences are the main driving forces of the amyloidogenicity of FapC (Rasmussen et al. 2019). Remarkably, a mutant FapC lacking all three amyloidogenic conserved imperfect repeats R1–R3 (and thus only consisting of N– and C–terminal tails, as well as the two linker regions), is still able to fibrillate but at very slow and stochastic rates and with a greatly increased tendency to fragment, showing a secondary role for the linker regions in the aggregation process. Actually, in our study, several residues residing in the linker regions are revealed, as Q227, N225, A223, E186, A187, and S189. This implies a possible role of the linker binding contacts of FapC with SPT in the impairment of the fibrilization process. These results suggest that the formation of the FapC fibril component/SPT protein complex in the extracellular space may possibly affect the FapC fibril biogenesis. As regards to the role of FapD is still a little unclear, though it has essential proteolytic activity necessary for FapC secretion (Rouse et al. 2017).
To summarize, the results of this study verify the anti-biofilm effect of the protease SPT—a natural-fermenting product with no adverse effects, against P. aeruginosa ATCC 27853, regarding both plastic and glass surfaces. The anti-biofilm effect of SPT was verified with the TCP method and by employing both optical and confocal microscopy. Additionally, this effect is interrelated with decreased titers of functional amyloids, as estimated by the employment of amyloid-specific dyes CR and ThT, and of lowered viability/metabolic activity of bacteria. In silico studies provided mechanistic details on the possible binding of FapC and FapD peptides by SPT and suggested a probable proteolytic mechanism for their clearance. Future studies should now be aiming for analyzing the effect of SPT on Pseudomonas amyloids and verify its possible proteolytic role and the exact binding mechanism on Fap peptides. Furthermore, a molecular explanation for viability inhibition based on studying various physiological pathways should be pursued. A well-organized in vivo study on lab animals with CF should also be attempted, in order to further elucidate the employment of SPT in clinical practice. Whatsoever, the present results will hopefully resurface the possible employment of SPT in medicine as a counter against bacterial biofilms and in combination with antibiotic therapy, to minimize the dose of the latter and thus stop the evolution of more multi-resistant strains – especially in healthcare facilities- and for the manifestation of chronic infections, like the ones in CF patients.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Al-Khateeb TH, Nusair Y (2008) Effect of the proteolytic enzyme serrapeptase on swelling, pain and trismus after surgical extraction of mandibular third molars. Int J Oral Maxillofac Surg 37:264–268. https://doi.org/10.1016/j.ijom.2007.11.011
Artini M, Papa R, Scoarughi GL, Galano E, Barbato G, Pucci P, Selan L (2013) Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J Appl Microbiol 114:266–277. https://doi.org/10.1111/jam.12038
Artini M, Scoarughi GL, Papa R, Cellini A, Carpentieri A, Pucci P, Amoresano A, Gazzola S, Cocconcelli PS, Selan L (2011) A new anti-infective strategy to reduce adhesion-mediated virulence in Staphylococcus aureus affecting surface proteins. Int J Immunopathol Pharmacol 24:661–672. https://doi.org/10.1177/039463201102400312
Artini M, Vrenna G, Trecca M, Tuccio Guarna Assanti V, Fiscarelli EV, Papa R, Selan L (2022) Serratiopeptidase affects the physiology of Pseudomonas aeruginosa isolates from cystic fibrosis patients. Int J Mol Sci 23:12645. https://doi.org/10.3390/ijms232012645
Bearden JC (1978) Quantitation of submicrogram quantities of protein by an improved protein-dye binding assay. Biochim Biophys Acta (BBA) - Protein Struct 533:525–529. https://doi.org/10.1016/0005-2795(78)90398-7
Beg AZ, Rashid F, Talat A, Haseen MA, Raza N, Akhtar K, Dueholm MKD, Khan AU (2023) Functional amyloids in Pseudomonas aeruginosa are essential for the proteome modulation that leads to pathoadaptation in pulmonary niches. Microbiol Spectr 11:e0307122. https://doi.org/10.1128/spectrum.03071-22
Bicking Kinsey C, Koirala S, Solomon B, Rosenberg J, Robinson BF, Neri A, Laufer Halpin A, Arduino MJ, Moulton-Meissner H, Noble-Wang J, Chea N, Gould CV (2017) Pseudomonas aeruginosa outbreak in a neonatal intensive care unit attributed to hospital tap water. Infect Control Hosp Epidemiol 38:801–808. https://doi.org/10.1017/ice.2017.87
Breathnach AS, Cubbon MD, Karunaharan RN, Pope CF, Planche TD (2012) Multidrug-resistant Pseudomonas aeruginosa outbreaks in two hospitals: association with contaminated hospital waste-water systems. J Hosp Infect 82:19–24. https://doi.org/10.1016/j.jhin.2012.06.007
Brown ML, Aldrich HC, Gauthier JJ (1995) Relationship between glycocalyx and povidone-iodine resistance in Pseudomonas aeruginosa (ATCC 27853) biofilms. Appl Environ Microbiol 61:187–193. https://doi.org/10.1128/aem.61.1.187-193.1995
Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJV, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI (2011) Tackling antibiotic resistance. Nat Rev Microbiol 9:894–896. https://doi.org/10.1038/nrmicro2693
Cao H, Lai Y, Bougouffa S, Xu Z, Yan A (2017) Comparative genome and transcriptome analysis reveals distinctive surface characteristics and unique physiological potentials of Pseudomonas aeruginosa ATCC 27853. BMC Genom 18:459. https://doi.org/10.1186/s12864-017-3842-z
Castelijn GAA, van der Veen S, Zwietering MH, Moezelaar R, Abee T (2012) Diversity in biofilm formation and production of curli fimbriae and cellulose of Salmonella Typhimurium strains of different origin in high and low nutrient medium. Biofouling 28:51–63. https://doi.org/10.1080/08927014.2011.648927
Chelliah V, Blundell TL, Fernández-Recio J (2006) Efficient restraints for protein-protein docking by comparison of observed amino acid substitution patterns with those predicted from local environment. J Mol Biol 357:1669–1682. https://doi.org/10.1016/j.jmb.2006.01.001
Chopra D, Rehan HS, Mehra P, Kakkar AK (2009) A randomized, double-blind, placebo-controlled study comparing the efficacy and safety of paracetamol, serratiopeptidase, ibuprofen and betamethasone using the dental impaction pain model. Int J Oral Maxillofac Surg 38:350–355. https://doi.org/10.1016/j.ijom.2008.12.013
Christensen LFB, Nowak JS, Sønderby TV, Frank SA, Otzen DE (2020) Quantitating denaturation by formic acid: imperfect repeats are essential to the stability of the functional amyloid protein FapC. J Biol Chem 295:13031–13046. https://doi.org/10.1074/jbc.RA120.013396
Dadgostar P (2019) Antimicrobial resistance: implications and costs. Infect Drug Resist 12:3903–3910. https://doi.org/10.2147/IDR.S234610
Danikowski KM, Cheng T (2018) Alkaline phosphatase activity of Staphylococcus aureus grown in biofilm and suspension cultures. Curr Microbiol 75:1226–1230. https://doi.org/10.1007/s00284-018-1514-0
Devlin H, Fulaz S, Hiebner DW, O’Gara JP, Casey E (2021) Enzyme-functionalized mesoporous silica nanoparticles to target Staphylococcus aureus and disperse biofilms. Int J Nanomedicine 16:1929–1942. https://doi.org/10.2147/IJN.S293190
Dueholm MS, Petersen SV, Sønderkær M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Otzen DE (2010) Functional amyloid in Pseudomonas. Mol Microbiol 77:1009–1020. https://doi.org/10.1111/j.1365-2958.2010.07269.x
Dueholm MS, Søndergaard MT, Nilsson M, Christiansen G, Stensballe A, Overgaard MT, Givskov M, Tolker-Nielsen T, Otzen DE, Nielsen PH (2013) Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2:365–382. https://doi.org/10.1002/mbo3.81
ECDC (2019) European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe 2018. Stockholm
Eisenberg D, Jucker M (2012) The amyloid state of proteins in human diseases. Cell 148:1188–1203. https://doi.org/10.1016/j.cell.2012.02.022
Fadl NN, Ahmed HH, Booles HF, Sayed AH (2013) Serrapeptase and nattokinase intervention for relieving Alzheimer’s disease pathophysiology in rat model. Hum Exp Toxicol 32:721–735. https://doi.org/10.1177/0960327112467040
Grela E, Kozłowska J, Grabowiecka A (2018) Current methodology of MTT assay in bacteria – a review. Acta Histochem 120:303–311. https://doi.org/10.1016/j.acthis.2018.03.007
Gupta PV, Nirwane AM, Belubbi T, Nagarsenker MS (2017) Pulmonary delivery of synergistic combination of fluoroquinolone antibiotic complemented with proteolytic enzyme: a novel antimicrobial and antibiofilm strategy. Nanomedicine 13:2371–2384. https://doi.org/10.1016/j.nano.2017.06.011
Hogan S, Zapotoczna M, Stevens NT, Humphreys H, O’Gara JP, O’Neill E (2017) Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections. J Hosp Infect 96:177–182. https://doi.org/10.1016/j.jhin.2017.02.008
Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O (2010) Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. https://doi.org/10.1016/j.ijantimicag.2009.12.011
Jadhav SB, Shah N, Rathi A, Rathi V, Rathi A (2020) Serratiopeptidase: Insights into the therapeutic applications. Biotechnol Rep 28:e00544. https://doi.org/10.1016/j.btre.2020.e00544
Jiménez-García B, Pons C, Fernández-Recio J (2013) pyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics 29:1698–1699. https://doi.org/10.1093/bioinformatics/btt262
Jones S (2017) Permeability rules for antibiotic design. Nat Biotechnol 35:639–639. https://doi.org/10.1038/nbt.3919
Katsipis G, Pantazaki AA (2023) Serrapeptase impairs biofilm, wall, and phospho-homeostasis of resistant and susceptible Staphylococcus aureus. Appl Microbiol Biotechnol 107:1373–1389. https://doi.org/10.1007/s00253-022-12356-5
Katsipis G, Tsalouxidou V, Halevas E, Geromichalou E, Geromichalos G, Pantazaki AA (2021) In vitro and in silico evaluation of the inhibitory effect of a curcumin-based oxovanadium (IV) complex on alkaline phosphatase activity and bacterial biofilm formation. Appl Microbiol Biotechnol 105:147–168. https://doi.org/10.1007/s00253-020-11004-0
Kv S, Devi GS, Mathew ST (2008) Liposomal formulations of serratiopeptidase: In vitro studies using PAMPA and Caco-2 models. Mol Pharm 5:92–97. https://doi.org/10.1021/mp700090r
Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH (2007) Amyloid adhesins are abundant in natural biofilms. Environ Microbiol 9:3077–3090. https://doi.org/10.1111/j.1462-2920.2007.01418.x
Leech J, Golub S, Allan W, Simmons MJH, Overton TW (2020) Non-pathogenic Escherichia coli biofilms: effects of growth conditions and surface properties on structure and curli gene expression. Arch Microbiol 202:1517–1527. https://doi.org/10.1007/s00203-020-01864-5
Longhi C, Scoarughi GL, Poggiali F, Cellini A, Carpentieri A, Seganti L, Pucci P, Amoresano A, Cocconcelli PS, Artini M, Costerton JW, Selan L (2008) Protease treatment affects both invasion ability and biofilm formation in Listeria monocytogenes. Microb Pathog 45:45–52. https://doi.org/10.1016/j.micpath.2008.01.007
Lyczak JB, Cannon CL, Pier GB (2002) Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. https://doi.org/10.1128/CMR.15.2.194-222.2002
Mecikoglu M, Saygi B, Yildirim Y, Karadag-Saygi E, Ramadan SS, Esemenli T (2006) The effect of proteolytic enzyme serratiopeptidase in the treatment of experimental implant-related infection. J Bone Joint Surg Am 88:1208–1214. https://doi.org/10.2106/JBJS.E.00007
Metkar SK, Girigoswami A, Murugesan R, Girigoswami K (2017) In vitro and in vivo insulin amyloid degradation mediated by Serratiopeptidase. Mater Sci Eng C, Mater Biol Appl 70:728–735. https://doi.org/10.1016/j.msec.2016.09.049
Metkar SK, Girigoswami A, Vijayashree R, Girigoswami K (2020) Attenuation of subcutaneous insulin induced amyloid mass in vivo using Lumbrokinase and Serratiopeptidase. Int J Biol Macromol 163:128–134. https://doi.org/10.1016/j.ijbiomac.2020.06.256
Mirani ZA, Fatima A, Urooj S, Aziz M, Khan MN, Abbas T (2018) Relationship of cell surface hydrophobicity with biofilm formation and growth rate: a study on Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli. Iran J Basic Med Sci 21:760–769. https://doi.org/10.22038/IJBMS.2018.28525.6917
Miyata K, Maejima K, Tomoda K, Isono M (1970) Serratia protease part I. Purification and general properties of the enzyme. Agric Biol Chem 34:310–318. https://doi.org/10.1080/00021369.1970.10859598
Moore NM, Flaws ML (2011) Epidemiology and pathogenesis of Pseudomonas aeruginosa infections. Clin Lab Sci 24:43–46
Nakamura S, Hashimoto Y, Mikami M, Yamanaka E, Soma T, Hino M, Azuma A, Kudoh S (2003) Effect of the proteolytic enzyme serrapeptase in patients with chronic airway disease. Respirology 8:316–320. https://doi.org/10.1046/j.1440-1843.2003.00482.x
Pachori P, Gothalwal R, Gandhi P (2019) Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis 6:109–119. https://doi.org/10.1016/j.gendis.2019.04.001
Papa R, Artini M, Cellini A, Tilotta M, Galano E, Pucci P, Amoresano A, Selan L (2013) A new anti-infective strategy to reduce the spreading of antibiotic resistance by the action on adhesion-mediated virulence factors in Staphylococcus aureus. Microb Pathog 63:44–53. https://doi.org/10.1016/j.micpath.2013.05.003
Passariello C, Lucchese A, Pera F, Gigola P (2012) Clinical, microbiological and inflammatory evidence of the efficacy of combination therapy including Serratiopeptidase in the treatment of periimplantitis. Eur J Inflamm 10:463–472. https://doi.org/10.1177/1721727X1201000322
Paytubi S, Cansado C, Madrid C, Balsalobre C (2017) Nutrient composition promotes switching between pellicle and bottom biofilm in Salmonella. Front Microbiol 8:2160. https://doi.org/10.3389/fmicb.2017.02160
Percival SL, Suleman L, Vuotto C, Donelli G (2015) Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol 64:323–334. https://doi.org/10.1099/jmm.0.000032
Pereira FDES, Bonatto CC, Lopes CAP, Pereira AL, Silva LP (2015) Use of MALDI-TOF mass spectrometry to analyze the molecular profile of Pseudomonas aeruginosa biofilms grown on glass and plastic surfaces. Microb Pathog 86:32–37. https://doi.org/10.1016/j.micpath.2015.07.005
Quick J, Cumley N, Wearn CM, Niebel M, Constantinidou C, Thomas CM, Pallen MJ, Moiemen NS, Bamford A, Oppenheim B, Loman NJ (2014) Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: an observational study using whole-genome sequencing. BMJ Open 4:e006278. https://doi.org/10.1136/bmjopen-2014-006278
Raman G, Avendano EE, Chan J, Merchant S, Puzniak L (2018) Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 7:79. https://doi.org/10.1186/s13756-018-0370-9
Rasmussen CB, Christiansen G, Vad BS, Lynggaard C, Enghild JJ, Andreasen M, Otzen D (2019) Imperfect repeats in the functional amyloid protein FapC reduce the tendency to fragment during fibrillation. Protein Sci 28:633–642. https://doi.org/10.1002/pro.3566
Rasmussen HØ, Kumar A, Shin B, Stylianou F, Sewell L, Xu Y, Otzen DE, Pedersen JS, Matthews SJ (2023) FapA is an intrinsically disordered chaperone for Pseudomonas functional amyloid FapC. J Mol Biol 435:167878. https://doi.org/10.1016/j.jmb.2022.167878
Reichhardt C, Jacobson AN, Maher MC, Uang J, McCrate OA, Eckart M, Cegelski L (2015) Congo red interactions with Curli-producing E. coli and native Curli amyloid fibers. PLoS One 10:e0140388. https://doi.org/10.1371/journal.pone.0140388
Reshamwala SMS, Noronha SB (2011) Biofilm formation in Escherichia coli cra mutants is impaired due to down-regulation of curli biosynthesis. Arch Microbiol 193:711–722. https://doi.org/10.1007/s00203-011-0708-7
Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, Molin S, Johansen HK (2021) Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 19:331–342. https://doi.org/10.1038/s41579-020-00477-5
Rouse SL, Hawthorne WJ, Berry J-L, Chorev DS, Ionescu SA, Lambert S, Stylianou F, Ewert W, Mackie U, Morgan RML, Otzen D, Herbst F-A, Nielsen PH, Dueholm M, Bayley H, Robinson CV, Hare S, Matthews S (2017) A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis. Nat Commun 8:263. https://doi.org/10.1038/s41467-017-00361-6
Rouse SL, Hawthorne WJ, Lambert S, Morgan ML, Hare SA, Matthews S (2016) Purification, crystallization and characterization of the Pseudomonas outer membrane protein FapF, a functional amyloid transporter. Acta Crystallogr F Struct Biol Commun 72:892–896. https://doi.org/10.1107/S2053230X16017921
Sannino G, Gigola P, Puttini M, Pera F, Passariello C (2013) Combination therapy including Serratiopeptidase improves outcomes of mechanical-antibiotic treatment of periimplantitis. Int J Immunopathol Pharmacol 26:825–831. https://doi.org/10.1177/039463201302600332
Selan L, Berlutti F, Passariello C, Comodi-Ballanti MR, Thaller MC (1993) Proteolytic enzymes: a new treatment strategy for prosthetic infections? Antimicrob Agents Chemother 37:2618–2621. https://doi.org/10.1128/AAC.37.12.2618
Selan L, Papa R, Ermocida A, Cellini A, Ettorre E, Vrenna G, Campoccia D, Montanaro L, Arciola CR, Artini M (2017) Serratiopeptidase reduces the invasion of osteoblasts by Staphylococcus aureus. Int J Immunopathol Pharmacol 30:423–428. https://doi.org/10.1177/0394632017745762
Selan L, Papa R, Tilotta M, Vrenna G, Carpentieri A, Amoresano A, Pucci P, Artini M (2015) Serratiopeptidase: a well-known metalloprotease with a new non-proteolytic activity against S. aureus biofilm. BMC Microbiol 15:207. https://doi.org/10.1186/s12866-015-0548-8
Shinde UA, Kanojiya SS (2014) Serratiopeptidase niosomal Gel with potential in topical delivery. J Pharm (Cairo) 2014:382959. https://doi.org/10.1155/2014/382959
Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR (2006) The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol 62:1264–1277. https://doi.org/10.1111/j.1365-2958.2006.05421.x
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. https://doi.org/10.1146/annurev.micro.56.012302.160705
Tachibana M, Mizukoshi O, Harada Y, Kawamoto K, Nakai Y (1984) A multi-centre, double-blind study of serrapeptase versus placebo in post-antrotomy buccal swelling. Pharmatherapeutica 3:526–530
Taglialegna A, Lasa I, Valle J (2016) Amyloid structures as biofilm matrix scaffolds. J Bacteriol 198:2579–2588. https://doi.org/10.1128/JB.00122-16
Thaller MC, Selan L, Fiorani P, Passariello C, Rizzo L, Speziale F (1997) A comparative in vitro evaluation of different therapeutic protocols for vascular graft infections. Eur J Vasc Endovasc Surg 14 Suppl A:35–37. https://doi.org/10.1016/s1078-5884(97)80151-7
Tsitsa I, Tarushi A, Doukoume P, Perdih F, De Almeida A, Papadopoulos A, Kalogiannis S, Casini A, Turel I, Psomas G (2016) Structure and biological activities of metal complexes of flumequine. RSC Adv 6:19555–19570. https://doi.org/10.1039/c5ra25776j
Van Gerven N, Van der Verren SE, Reiter DM, Remaut H (2018) The role of functional amyloids in bacterial virulence. J Mol Biol 430:3657–3684. https://doi.org/10.1016/j.jmb.2018.07.010
Vélez-Gómez JM, Melchor-Moncada JJ, Veloza LA, Sepúlveda-Arias JC (2019) Purification and characterization of a metalloprotease produced by the C8 isolate of Serratia marcescens using silkworm pupae or casein as a protein source. Int J Biol Macromol 135:97–105. https://doi.org/10.1016/j.ijbiomac.2019.05.122
Wang H, Cheng H, Wang F, Wei D, Wang X (2010) An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J Microbiol Methods 82:330–333. https://doi.org/10.1016/j.mimet.2010.06.014
Yang L, Haagensen JAJ, Jelsbak L, Johansen HK, Sternberg C, Høiby N, Molin S (2008) In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J Bacteriol 190:2767–2776. https://doi.org/10.1128/JB.01581-07
Yin W, Wang Y, Liu L, He J (2019) Biofilms: The microbial “protective clothing” in extreme environments. Int J Mol Sci 20:3423. https://doi.org/10.3390/ijms20143423
Zapotoczna M, McCarthy H, Rudkin JK, O’Gara JP, O’Neill E (2015) An essential role for Coagulase in Staphylococcus aureus biofilm development reveals new therapeutic possibilities for device-related infections. J Infect Dis 212:1883–1893. https://doi.org/10.1093/infdis/jiv319
Zeng G, Vad BS, Dueholm MS, Christiansen G, Nilsson M, Tolker-Nielsen T, Nielsen PH, Meyer RL, Otzen DE (2015) Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front Microbiol 6:1099. https://doi.org/10.3389/fmicb.2015.01099
Zor T, Selinger Z (1996) Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem 236:302–308. https://doi.org/10.1006/abio.1996.0171
Acknowledgements
The authors would like to acknowledge the Laboratory of Engineering Mechanics of the Civil Engineering Department of AUTh for access to the Large Research Infrastructure and Instrumentation of the Laboratory, purchased through MIS5010629.
Funding
Open access funding provided by HEAL-Link Greece.
Author information
Authors and Affiliations
Contributions
GK and AP conceptualized the study. GK conducted the microbiological and biochemical experiments, performed the statistical analysis, and wrote the first draft of the manuscript. GK and DA performed the fluorescence microscopy experiments, analyzed data, and wrote the corresponding manuscript sections. MP and DA performed the confocal microscopy experiments, analyzed data, and wrote the corresponding manuscript sections. Molecular docking studies were carried out by GG, who also wrote the corresponding manuscript sections. AP and MP provided the needed laboratory equipment and materials. MP supervised all microscopy examinations. AP coordinated the study, supervised all microbiological and biochemical experiments, and corrected the first draft of the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Katsipis, G., Avgoulas, D.I., Geromichalos, G.D. et al. In vitro and in silico evaluation of the serrapeptase effect on biofilm and amyloids of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 107, 7269–7285 (2023). https://doi.org/10.1007/s00253-023-12772-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12772-1