Abstract
Background
Identifying risk factors predicting acquisition of resistant Pseudomonas aeruginosa will aid surveillance and diagnostic initiatives and can be crucial in early and appropriate antibiotic therapy. We conducted a systematic review examining risk factors of acquisition of resistant P. aeruginosa among hospitalized patients.
Methods
MEDLINE®, EMBASE®, and Cochrane Central were searched between 2000 and 2016 for studies examining independent risk factors associated with acquisition of resistant P. aeruginosa, among hospitalized patients. Random effects model meta-analysis was conducted when at least three or more studies were sufficiently similar.
Results
Of the 54 eligible articles, 28 publications (31studies) examined multi-drug resistant (MDR) or extensively drug resistant (XDR) P. aeruginosa and 26 publications (29 studies) examined resistant P. aeruginosa. The acquisition of MDR P. aeruginosa, as compared with non-MDR P. aeruginosa, was significantly associated with intensive care unit (ICU) admission (3 studies: summary adjusted odds ratio [OR] 2.2) or use of quinolones (4 studies: summary adjusted OR 3.59). Acquisition of MDR or XDR compared with susceptible P. aeruginosa was significantly associated with prior hospital stay (4 studies: summary adjusted OR 1.90), use of quinolones (3 studies: summary adjusted OR 4.34), or use of carbapenems (3 studies: summary adjusted OR 13.68). The acquisition of MDR P. aeruginosa compared with non-P. aeruginosa was significantly associated with prior use of cephalosporins (3 studies: summary adjusted OR 3.96), quinolones (4 studies: summary adjusted OR 2.96), carbapenems (6 studies: summary adjusted OR 2.61), and prior hospital stay (4 studies: summary adjusted OR 1.74). The acquisition of carbapenem-resistant P. aeruginosa compared with susceptible P. aeruginosa, was statistically significantly associated with prior use of piperacillin-tazobactam (3 studies: summary adjusted OR 2.64), vancomycin (3 studies: summary adjusted OR 1.76), and carbapenems (7 studies: summary adjusted OR 4.36).
Conclusions
Prior use of antibiotics and prior hospital or ICU stay was the most significant risk factors for acquisition of resistant P. aeruginosa. These findings provide guidance in identifying patients that may be at an elevated risk for a resistant infection and emphasize the importance of antimicrobial stewardship and infection control in hospitals.
Similar content being viewed by others
Background
There is an alarming increase in antibiotic-resistant Gram-negative infections [1–3]. Among Gram-negative infections, Pseudomonas aeruginosais is one of the most common gram-negative bacteria causing nosocomial and healthcare-associated infections (HAIs) in hospitalized patients [4]. The World Health Organization places carbapenem-resistant P. aeruginosa as a critical priority pathogen that desperately requires new treatment options [5]. Increasing rates of multidrug-resistant (MDR) P. aeruginosa in HAIs and among hospitalized patients is a major public health problem [6]. MDR P. aeruginosa infections in the hospital setting are associated with poor outcomes including increased resource utilization and costs, morbidity, and mortality [7].
MDR P. aeruginosa account for 13–19% of HAIs each year in the US. The increasing level of resistance in MDR P. aeruginosa is often attributed to patient-to-patient transmission of resistant strains as well as newly acquired resistance owing to previous antibiotic exposure. As per standardized international terminology, MDR is defined as non-susceptibility to at least one agent in three or more antimicrobial categories and XDR is defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e. bacterial isolates remain susceptible to only one or two categories) [8]. In severe systemic infections, consideration of MDR/XDR P. aeruginosa when selecting treatment is warranted to ensure timely and appropriate initial therapy. Lack of effective antibacterial therapies against P. aeruginosa infections severely limit effective therapeutic options and consequently, lead to inappropriate initial therapy that adversely impacts health outcomes [9]. Typically it takes 48 h to definitively identify MDR P. aeruginosa, which can be a detrimental delay for these patients.
There are considerable gaps and inconsistencies in knowledge regarding risk factors associated with the occurrence of MDR P. aeruginosa in nosocomial infections and HAIs. Identifying risk factors predicting acquisition of MDR P. aeruginosa or identifying subgroups of patients who are at an increased risk for acquisition of MDR P. aeruginosa in the hospital setting will assist in providing timely and appropriate treatment. While a recent review examined the risk factors independently associated with extensively drug-resistant (XDR) P. aeruginosa, there has not been a comprehensive analysis of contemporary literature reporting all levels of resistance (MDR or XDR or resistant) of P. aeruginosa infections [6]. This systematic review of recent published literature evaluates risk factors that are independently associated with acquisition of microbiologically identified MDR or XDR, or resistant (single drug class) P. aeruginosa from various sites among inpatients, hospitalized in any of the following setting including wards, intensive care units, and other types of inpatient settings.
Methods
We performed a systematic search in the MEDLINE, Cochrane Library, and EMBASE databases for citations indexed from January 01, 2000 through December 31, 2016. The initial search strategy (Additional file 1: Table S1) includes terms related to the pathogen (P. aeruginosa), mode of infection (nosocomial, hospital-acquired, healthcare-acquired, hospital-associated, healthcare-associated), and risk factor assessment (risk factors, predict, risk score, risk assessment, and multivariate analysis). Additionally, search terms for Gram-negative infections, beta-lactamases, and metallo-beta-lactamases attributed to carbapenem resistance were included to increase the yield of studies with resistant (single drug class) P. aeruginosa. These three databases were searched because they index most of the published citations. To supplement this search, we reviewed reference lists of eligible studies using an iterative process to maximize inclusion of relevant data. We did not perform grey literature searches or systematically search for unpublished data. We reviewed abstracts from conference proceedings if they were indexed in the three aforementioned databases.
Study definitions
Our primary objective was to examine studies evaluating risk factors associated with acquisition of MDR or XDR, or resistant P. aeruginosa among inpatient adult patients evaluated in the hospital setting. In this review, the hospital setting comprises all types of units, including intensive care units, emergency room/casualty, or other wards where patients were located at the time of collection of the resistant P. aeruginosa microbiological specimen and underwent further treatment in the hospital setting. The acquisition of resistant P. aeruginosa could have been either community- or hospital-acquired, but all patients must have been evaluated and treated in the hospital setting. We accepted MDR and XDR P. aeruginosa as defined by individual studies, although we noted considerable heterogeneity in the definitions. Non-MDR P. aeruginosa was also defined by the individual studies and varied in definition to include P. aeruginosa infections other than MDR such as susceptible P. aeruginosa (susceptible to all antipseudomonal agents) or resistant to any one class of drug. Non- P. aeruginosa was also defined by the individual studies and varied in definition to include pathogens other than P. aeruginosa.
Eligibility criteria
Studies evaluating patients hospitalized in a ward, intensive or critical care unit, or any other inpatient healthcare setting (including chronic care facilities), were eligible for inclusion. Studies examining patients with either hospital-acquired or community-acquired were included, provided that they were evaluated in a hospital /inpatient setting. Risk factors that predict acquisition of MDR or XDR, or resistant P. aeruginosa were included. Risk factors were categorized into patient characteristics, hospital characteristics, and treatment characteristics.
The exposure and comparators of interest included: MDR or XDR P. aeruginosa versus resistant P. aeruginosa (to any one class of antibiotic); MDR or XDR P. aeruginosa versus susceptible P. aeruginosa; MDR or XDR P. aeruginosa versus control (any other Gram-negative pathogen or any infection with unspecified pathogen); resistant P. aeruginosa versus susceptible P. aeruginosa. The outcome of interest was microbiologically confirmed acquisition of MDR or XDR, or resistant P. aeruginosa. For studies that reported both the outcome of acquisition of MDR or XDR P. aeruginosa and single-drug resistant P. aeruginosa, we included the study only once (for the outcome of acquisition of MDR or XDR P. aeruginosa) in the analysis. We accepted all study designs except non-comparative studies, case reports, and case series. Studies that reported multivariable results adjusting for any potential confounders were eligible. No minimum study duration or follow-up time was required for inclusion.
We excluded studies of mixed infections that had more than 20% Gram-positive infections (to ensure most (80%) of the evaluated infections in eligible studies were Gram-negative infections) and studies solely examining P. aeruginosa resistant due to metallo-beta-lactamases. Studies published in languages other than English were excluded.
All citations identified by literature searches were independently screened by two researchers. Upon the start of citation screening, we implemented a training session where all researchers screened the same articles and conflicts were discussed. We iteratively continued training until we reached consensus regarding the nuances of the eligibility criteria for screening. During double-screening, we resolved conflicts through group discussions.
We assessed the methodological quality of each study based on predefined criteria using the Agency for Healthcare Research and Quality (AHRQ) and the New-Castle Ottawa risk of bias tool, which probes risk of selection, performance, detection, attrition, reporting, and other potential biases. Each study was extracted by one experienced methodologist (GR, EA, and JC). The extraction was reviewed and confirmed by at least one other methodologist. Any disagreements were resolved by discussion amongst the study team. Data were extracted into customized Microsoft Excel™ forms. We tested the data extraction forms on several studies and revised as necessary before full data extraction. Extracted data included variables addressing population characteristics, including severity of illness, description of exposure and comparator groups, sites of infection, outcome definitions, enrolled and analyzed sample sizes, study design features, and multivariate results. Any missing or unavailable data were deemed as not reported information. We performed random effects model meta-analyses of eligible studies if at least three or more studies were sufficiently similar in population, exposure, and outcomes [10]. If appropriate, we also conducted meta-regression analyses to evaluate study features explaining potential heterogeneity. However, due to the presence of substantial clinical heterogeneity, we also qualitatively compared results across studies (for example, source of infection).
All analyses were performed in Stata version 14 (StataCorp, College Station, Texas) with the metan, metareg, and metabias functions. We tested between study heterogeneity with the Q statistic (significant when p < 0.10) and quantified the extent of heterogeneity with the I2 statistic. It is common practice to interpret I2 > 50% as representing substantial inconsistency or significant statistical heterogeneity [11].
When applicable, results were also presented in table format. All meta-analyses of adjusted or multivariate results were presented in forest plots. The summary results examining the relationship between risk factors and acquisition of MDR or XDR, or resistant P. aeruginosa were presented as adjusted odds ratio or relative risks with 95% confidence intervals.
Results
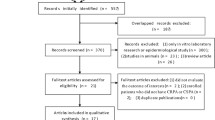
The literature search identified 316 citations and an additional 43 citations were identified through bibliographic searches. After removing duplicates, 345 abstracts were eligible for screening. Using a low threshold of eligibility criteria for review of abstracts, 112 full-text articles were retrieved. After full-text review, an additional 58 articles were excluded based on the eligibility criteria (Fig. 1). Of the 54 eligible articles, 28 articles (31 studies) examined MDR or XDR P. aeruginosa [12–39] and the remaining 26 articles (29 studies) examined resistant P. aeruginosa [41–65].
Studies of MDR and XDR P. aeruginosa
Of the 28 articles (n = 8935 subjects) that examined risk factors for acquisition of MDR or XDR P. aeruginosa [12–39], nine were prospective studies and 19 were retrospective or unmatched case-control or matched case-control studies. In 31 studies from 28 articles; two articles reported two different control groups [20, 27] and one study reported two different cases [33]. The studies were conducted in Western European countries (16 studies), the USA (3 studies), the USA and Western Europe (2 studies), Asia (6 studies), Turkey (1 study), or Brazil (1 study); 3 studies were conducted in an unclear location (Table 1). Of the 28 articles, three were multi-center studies [26, 32, 35]. Twenty-one were conducted in tertiary care or university hospitals and the other 9 were conducted in any hospital.
Of the included 28 articles, 20 articles (from 22 studies) reported data on MDRP. aeruginosa acquisition [12, 14–20, 22, 23, 25–31, 35, 37, 39], six reported on XDR P. aeruginosa [13, 24, 32–34, 38], and two included both (MDR acquisition being the most commonly reported) [21, 36]. The most commonly reported definition for MDR was laboratory-confirmed resistance to more than one agent in three or more classes of antibiotics. In five studies, the definitions were similar to the definition of XDR, but they were described as MDR [12, 16, 17, 27, 28]. The most commonly reported definition for XDR was non-susceptibility to at least one agent in all antimicrobial classes, but susceptibility to two or fewer anti-pseudomonal antimicrobials; one study required evidence of resistance to all available anti-pseudomonal antimicrobials [32].
Of 8935 patients, 2446 case patients had MDR or XDR P. aeruginosa or both types of resistance and the remaining were control patients. The descriptions of study control groups varied across studies: susceptible P. aeruginosa (5 studies [12, 13, 26, 29, 32]); non-MDR P. aeruginosa (without MDR phenotype) (14 studies [14, 15, 18–22, 24, 25, 27, 34, 35, 60, 39]); non-XDR P. aeruginosa (without XDR phenotype) (2 studies [31, 33]); non-P. aeruginosa (any pathogen other than P. aeruginosa) (4 studies [16, 23, 26, 30]); non-MDR/non-XDR P. aeruginosa plus non-P. aeruginosa (2 studies [17, 37]). No data were reported in two studies [11, 19]. The mean age of study subjects ranged from 29.8 years to 73.2 years (cases) and from 37.9 years to 71.1 years (controls). The males included as study subjects ranged from 50 to 91% (cases) and from 46 to 78.3% (controls). Most studies included a mix of patients with respiratory, wound, genitourinary, and bloodstream as the source of infection. Various severity scores were employed across studies including McCabe score, APACHE II, Charlson index, and SAPS II score.
For meta-analysis, we categorized studies based on three control groups: non-MDR P. aeruginosa (without MDR phenotype); susceptible P. aeruginosa; and any controls (non P. aeruginosa or undescribed controls). In analyses of risk for acquisition of MDR P. aeruginosa, 21 articles in 23 studies [12, 14–20, 22, 23, 25–31, 33, 35, 37, 39]) reported data on MDR P. aeruginosa acquisition and two included both MDR P. aeruginosa and XDR P. aeruginosa (MDR P. aeruginosa acquisition being the most commonly reported) [21, 36].
The risk factors statistically significantly associated with acquisition of MDR P. aeruginosa in meta-analysis are described in the following results sections. Additional risk factors that had a statistically significant association with acquisition of MDR P. aeruginosa but were not eligible for meta-analysis are listed in Additional file 1: Tables S2-S5.
MDR P. aeruginosa versus non-MDR P. aeruginosa
Thirteen studies with non-MDR P. aeruginosa as a comparator reported risk factors associated with acquisition of MDR P. aeruginosa [15, 16, 19–23, 25, 26, 28, 35, 36, 39]. In meta-analysis, prior ICU stay or prior use of quinolones had a statistically significant association with acquisition of MDR P. aeruginosa. Two other predictors, surgery and prior use of carbapenems, were not associated with an increased risk of acquisition (Fig. 2).
MDR P. aeruginosa versus susceptible P. aeruginosa
Four studies with susceptible P. aeruginosa as a comparator reported risk factors for acquisition of MDR P. aeruginosa [14, 27, 30, 33]. In meta-analysis, prior admission, use of quinolones, and use of carbapenems had a statistically significant association with future acquisition of MDR P. aeruginosa. Two other predictors (comorbid severity scores and chronic obstructive pulmonary disease [COPD]) were not associated with a risk of acquisition of MDR P. aeruginosa in meta-analysis (Fig. 3).
Of note, the following factors were not combined in a meta-analysis; but were reported to be not associated with acquisition of MDR P. aeruginosa: colonization of P. aeruginosa in isolate sources other than blood preceding a diagnosis of bacteremia, isolation of multiple pathogens in culture, and history of previous P. aeruginosa infection.
MDR P. aeruginosa versus non-P. aeruginosa
Seven studies with non-P. aeruginosa or any undescribed control as a comparator reported risk factors for acquisition of MDR P. aeruginosa [12, 17, 18, 27, 29, 31, 37]. In meta-analysis, penicillins, carbapenems, quinolones, disease severity, and prior admissions predicted a statistically significant increased risk of acquiring MDR P. aeruginosa, compared with non-P. aeruginosa (Fig. 4).
Both diabetes and sepsis were not associated with the risk of acquiring MDR P. aeruginosa, compared with non-P. aeruginosa.
XDR P. aeruginosa studies
Six studies reported risk factors for acquisition of XDR P. aeruginosa alone, compared with different controls [13, 24, 32–34, 38]. XDR P. aeruginosa was compared with non-XDR P. aeruginosa (2 studies [32, 34]); non-XDR P. aeruginosa and non-P. aeruginosa (1 study [38]); non-P. aeruginosa (1 study [24]); susceptible P. aeruginosa (1 study [33]); and susceptible P. aeruginosa and non-P. aeruginosa (1 study [13]).The most frequently reported factors that significantly increased the risk of acquiring XDR P. aeruginosa included: prior fluoroquinolones use (2 studies), urinary catheter (2 studies). The following factors were not associated with an increased risk of XDR P. aeruginosa: ceftazidime use, days of ciprofloxacin use, number of different antibiotics used during time at risk (i.e. time between admission and isolation of bacteria in index cases).
Studies of resistant P. aeruginosa
Carbapenem-resistant P. aeruginosa
Twenty-three studies in 21 articles (n = 9877 subjects) that examined risk factors for acquisition of carbapenem-resistant P. aeruginosa [17, 19, 40, 42–47, 50, 51, 54–57, 59, 60, 61, 62, 64, 65]; two articles contributed to two studies each [44, 64]. Five were prospective studies and the remaining 16 had case-control or retrospective designs. The studies were conducted in Brazil (8 studies), the USA (5 studies), Western Europe (4 studies), Asia (3 studies), and the USA and Western Europe (1 study). All studies were conducted in tertiary care or university hospitals. Three were multicenter studies. Table 2 summarizes baseline characteristics of included carbapenem resistant P. aeruginosa studies.
The descriptions of control groups varied across studies. For the purpose of analysis, we categorized the controls into two categories: susceptible P. aeruginosa and any control (non-P. aeruginosa with or without susceptible P. aeruginosa). The mean age ranged from 44 years to 67 years (cases) and from 41 years to 66 years (controls). Most studies included a mix of patients with respiratory, wound, genitourinary, and bloodstream as the source of infection.
Carbapenem-resistant versus susceptible P. aeruginosa
In meta-analysis, prior use of piperacillin-tazobactam, vancomycin, and carbapenems were all significantly associated with acquisition of carbapenem-resistant P. aeruginosa (Additional file 1: Figures S1-S2). Other predictors including use of quinolones, hospital stay, and time at risk were not associated with a statistically significant increased risk (Additional file 1: Figures S1-S2). Additional risk factors that significantly increased the risk of acquiring carbapenem-resistant P. aeruginosa are summarized in Additional file 1: Tables S6-S9.
Carbapenem-resistant versus any control (unspecified or non- P. aeruginosa controls)
Four studies used other controls (3 studies that used Non-P.aeruginosa controls and one study did not specify the control) [17, 44, 45, 64]. No meta-analysis could be conducted as there were fewer than three studies for this comparison for the same type of predictor. All studies reported a statistically significant increased risk of carbapenem-resistant P. aeruginosa, compared with any control. The risk factors examined included prior use of antibiotics, comorbid score, length of hospital or ICU stay, hemodialysis, non-ambulatory status, transfer from another facility, indwelling urinary catheter, and mechanical ventilation.
Other (non-carbapenem) resistant P. aeruginosa
Of the seven studies that reported data on acquisition of other (non-carbapenem) resistant P. aeruginosa in Table 3 [41, 45, 48, 49, 52, 53, 63], two reported data on piperacillin- or piperacillin-tazobactam-resistant P. aeruginosa [49, 63], three reported data on quinolone-resistant P. aeruginosa [48, 52, 53], and two reported on P. aeruginosa resistant to the newer cephalosporins, namely, cefepime and ceftazidime [41, 45].
Piperacillin- or piperacillin-tazobactam-resistant P. aeruginosa
No meta-analysis could be conducted as there were fewer than three studies for this comparison on any particular category of predictor [49, 63]. Two studies reported a statistically significant increased risk for piperacillin- or piperacillin-tazobactam-resistant P. aeruginosa for the following factors: admissions in prior year, ICU stay, time at risk, transfer, severe morbidity or higher comorbid score, and prior use of antibiotics (aminoglycosides, broad-spectrum cephalosporins, carbapenems, piperacillin-tazobactam, and quinolones).
Quinolone-resistant P. aeruginosa
Three studies compared quinolone-resistant and susceptible P. aeruginosa [48, 53, 53]. In meta-analysis, prior use of quinolones predicted subsequent risk for acquisition of quinolone-resistant P. aeruginosa (Additional file 1: Figure S3). Other statistically significant risk factors for an increased acquisition of quinolone-resistant P. aeruginosa included indwelling airway, co-existing diabetes mellitus, and nosocomial residence (not defined) (Additional file 1: Table S10). The number of hospital days from admission to culture was not a significant predictor.
Third-generation cephalosporin-resistant P. aeruginosa
Two studies [41, 45] reported an increased risk for third-generation cephalosporin-resistant P. aeruginosa for the following factors: transfer from outside facility, prior use of antibiotics (e.g. extended-spectrum cephalosporins, extended-spectrum penicillin, or quinolones or amikacin). Both length of hospital stay before culture and prior use of carbapenem were not associated with an increased risk for resistance to higher generation cephalosporins in one study [41].
Discussion
This systematic review of the literature summarizes risk factors that predict acquisition of MDR P. aeruginosa, XDR P. aeruginosa, and other resistant P. aeruginosa. Our meta-analysis identified that the risk factors for acquisition of MDR or XDR P. aeruginosa varied with regard to the type of control used for comparison. ICU stays (either as current or prior to current episode of infection) and use of quinolones was significantly associated with acquisition of MDR or XDR P. aeruginosa, compared with resistant or susceptible P. aeruginosa. Use of prior antibiotics including, cephalosporins, quinolones, or carbapenems, and prior hospital admissions predicted an increased risk of MDR or XDR P. aeruginosa versus non-P. aeruginosa.
The increasing rates of MDR or XDR, or resistant P. aeruginosa are a worldwide public health problem. Resistant strains of P. aeruginosa are associated with high mortality and increased resource utilization [66]. The emergence and spread of MDR P. aeruginosa may be associated with misuse or overuse of antimicrobials. Our meta-analysis found a consistent association between the use of quinolones and acquisition of MDR or XDR P. aeruginosa P. aeruginosa. However, association between carbapenem use and acquisition of MDR or XDR P. aeruginosa was not consistent across comparisons. While some primary studies did report a relationship between MDR or XDR P. aeruginosa and prior use of antibiotics [14, 30], our meta-analysis identified statistically significant associations between carbapenem use and acquisition of MDR or XDR P. aeruginosa and the significance of their association can vary by the type of comparator examined. Nonetheless, our results confirm that prior use of specific antimicrobials is an important risk factor for acquisition of MDR or XDR P. aeruginosa. Antimicrobial resistance occurs over time, often mediated by gene mutations and misuse or overuse of antimicrobials may accelerate this process. Understanding the epidemiology of MDR or XDR P. aeruginosa will be necessary to overcome infections with these resistant pathogens.
This review finding can be supported in the context of a recent review on MDR or XDR P. aeruginosa, which examined multivariate risk factors reported in eight included articles [67]. The authors of this review did not perform a meta-analysis but identified prior antimicrobial therapy, medical devices, patient-related characteristics, and environmental sources as risk factors for MDR or XDR P. aeruginosa. Our review, in which a meta-analysis was performed, demonstrated that admission location, prior admission, or use of quinolones was significantly associated with acquisition of MDR or XDR P. aeruginosa. In addition to reviewing MDR or XDR P. aeruginosa, we reviewed single drug-resistant P. aeruginosa. In meta-analysis, prior use of piperacillin-tazobactam, vancomycin, or carbapenems were significantly associated with acquisition of carbapenem-resistant versus susceptible P. aeruginosa. In meta-analysis, prior fluoroquinolone use was a statistically significant predictor of subsequent quinolone-resistant versus susceptible P. aeruginosa.
There are limitations associated with this systematic literature review. To begin with, the study inclusion criteria were limited to published studies indexed in English. Secondly, studies eligible for inclusion were heterogeneous with respect to the definition of exposure, site of infection with P. aeruginosa, and risk factors included in the study. As a result, a combined meta-analysis may misrepresent the true picture. We mitigated these issues by using a random effects model in our analyses and by limiting analyses to similar comparators. However, in many instances there were risk factors that were statistically significant in two studies, but unfortunately a third study was not available to permit the exploration of this factor through meta-analysis. Additionally, our study results may not be generalizable to all regions as studies from certain regions (for example, Western European nations for MDR or XDR P. aeruginosa and Brazil for carbapenem-resistant P. aeruginosa) were overrepresented in our sample published literature.
Confounding in observational studies reporting unadjusted data is a well-known source of bias. To mitigate this bias, we included risk factors that were analyzed in multivariate analyses. The choice of the control group (e.g. patients with infection with susceptible strains or patients without any infection at all) may also have influenced the results. We attempted to address this issue using similar comparison groups in the meta-analyses. As with any evidence synthesis, the limitations of the data available in primary studies will transfer into limitations of the systematic review. For example, the control group was not well defined in studies, specifically in studies that reported non- P. aeruginosa as controls. The description of these controls was unclear if the control group did or did not include MDR infection of a non-Pseudomonas bacteria. The results section describes variability across studies with regard to study characteristics, outcome assessment, and the relationship between risk factors and the outcome of acquisition of resistant P. aeruginosa. We could not conduct additional subgroup analyses by site of infection, or explain differences across studies using stratified analyses due to the small number of available studies for any particular comparison. An insufficient number of studies (< 10 studies) precluded evaluatingthe potential for publication bias with funnel plots and Egger’s tests for small study effects [68].
Conclusions
Consistently, across comparisons, prior use of antibiotics and prior ICU stay was a significant risk predictor for acquisition of MDR or XDR P. aeruginosa infections. Depending on local epidemiology, this finding is useful in identifying patients at high risk for resistant P. aeruginosa that may benefit from alternate empiric treatment. Further, these findings emphasize the need for antimicrobial stewardship and infection control in hospitals and continued need for the development of new antimicrobial agents with activity against MDR P. aeruginosa [68]. The implementation of antimicrobial stewardship and infection control in hospitals can improve patient safety and care, reduce resource utilization, and reduce resistance. The increasing prevalence of antimicrobial resistance among hospitalized patients continues to pose a challenge for practitioners.
Abbreviations
- AHRQ:
-
Agency for Healthcare Research and Quality
- APACHE II:
-
Acute Physiology and Chronic Health Evaluation II
- COPD:
-
Chronic obstructive pulmonary disease
- ESBL:
-
Extended-spectrum beta-lactamase organisms
- HAI:
-
Hospital acquired infection
- ICU:
-
Intensive care unit
- MDR:
-
Multi-drug resistant
- OR:
-
Odds ratio
- SAPS II:
-
Simplified acute physiology score II
- US:
-
United States
- XDR:
-
Extensively drug resistant
References
Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1):1–12.
Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362(19):1804–13.
Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302(21):2323–9.
Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3):351–68.
Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ 2017 February 27 [cited 2017 Dec 5];
Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48(12):4606–10.
Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3(1):32.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Raman G, Avendano E, Berger S, Menon V. Appropriate initial antibiotic therapy in hospitalized patients with gram-negative infections: systematic review and meta-analysis. BMC Infect Dis. 2015;15:395.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50(1):43–8.
Bodro M, Sabe N, Tubau F, Llado L, Baliellas C, Gonzalez-Costello J, et al. Extensively drug-resistant Pseudomonas aeruginosa bacteremia in solid organ transplant recipients. Transplantation. 2015;99(3):616–22.
Cao B, Wang H, Sun H, Zhu Y, Chen M. Risk factors and clinical outcomes of nosocomial multi-drug resistant Pseudomonas aeruginosa infections. J Hosp Infect. 2004;57(2):112–8.
Cilloniz C, Gabarrus A, Ferrer M, Puig dB, Rinaudo M, Mensa J, et al. Community-acquired pneumonia due to multidrug- and non-multidrug-resistant Pseudomonas aeruginosa. Chest. 2016;150(2):415–25.
Cobos-Trigueros N, Sole M, Castro P, Torres JL, Hernandez C, Rinaudo M, et al. Acquisition of Pseudomonas aeruginosa and its resistance phenotypes in critically ill medical patients: role of colonization pressure and antibiotic exposure. Crit Care. 2015;19:218.
D'Agata EM, Cataldo MA, Cauda R, Tacconelli E. The importance of addressing multidrug resistance and not assuming single-drug resistance in case-control studies. Infect Control Hosp Epidemiol. 2006;27(7):670–4.
Dalfino L, Mosca A, Brienza N, Spada M, Coppolecchia S, Miragliotta G, et al. Nosocomial infections in critically ill patients: When should multidrug-resistant Pseudomonas aeruginosa be suspected? Clin Microbiol Infect. 2011;17:S294–5. Conference: 21st ECCMID/27th ICC Milan Italy Conference Start: 20110507 Conference End: 20110510 Conference Publication: (var pagings)
Dantas RC, Ferreira ML, Gontijo-Filho PP, Ribas RM. Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol. 2014;63(Pt:12):12–87.
Defez C, Fabbro-Peray P, Bouziges N, Gouby A, Mahamat A, Daures JP, et al. Risk factors for multidrug-resistant Pseudomonas aeruginosa nosocomial infection. J Hosp Infect. 2004;57(3):209–16.
Gomez-Zorrilla S, Camoez M, Tubau F, Periche E, Canizares R, Dominguez MA, et al. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob Agents Chemother. 2014;58(10):5863–70.
Johnson LE, D'Agata EM, Paterson DL, Clarke L, Qureshi ZA, Potoski BA, et al. Pseudomonas aeruginosa bacteremia over a 10-year period: multidrug resistance and outcomes in transplant recipients. Transpl Infect Dis. 2009;11(3):227–34.
Joo EJ, Kang CI, Ha YE, Kang SJ, Park SY, Chung DR, et al. Risk factors for mortality in patients with Pseudomonas aeruginosa bacteremia: clinical impact of antimicrobial resistance on outcome. Microb Drug Resist. 2011;17(2):305–12.
Liew YX, Tan TT, Lee W, Ng JL, Chia DQ, Wong GC, et al. Risk factors for extreme-drug resistant Pseudomonas aeruginosa infections in patients with hematologic malignancies. Am J Infect Control. 2013;41(2):140–4.
Lodise TP, Miller CD, Graves J, Furuno JP, McGregor JC, Lomaestro B, et al. Clinical prediction tool to identify patients with Pseudomonas aeruginosa respiratory tract infections at greatest risk for multidrug resistance. Antimicrob Agents Chemother. 2007;51(2):417–22.
Micek ST, Wunderink RG, Kollef MH, Chen C, Rello J, Chastre J, et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Critical Care (London, England). 2015;19:219.
Montero M, Sala M, Riu M, Belvis F, Salvado M, Grau S, et al. Risk factors for multidrug-resistant Pseudomonas aeruginosa acquisition. Impact of antibiotic use in a double case-control study. Eur J Clin Microbiol Infect Dis. 2010;29(3):335–9.
Nakamura A, Miyake K, Misawa S, Kuno Y, Horii T, Kondo S, et al. Meropenem as predictive risk factor for isolation of multidrug-resistant Pseudomonas aeruginosa. J Hosp Infect. 2013;83(2):153–5.
Nseir S, Blazejewski C, Lubret R, Wallet F, Courcol R, Durocher A. Risk of acquiring multidrug-resistant gram-negative bacilli from prior room occupants in the intensive care unit. Clin Microbiol Infect. 2011;17(8):1201–8.
Ohmagari N, Hanna H, Graviss L, Hackett B, Perego C, Gonzalez V, et al. Risk factors for infections with multidrug-resistant Pseudomonas aeruginosa in patients with cancer. Cancer. 2005;104(1):205–12.
Paramythiotou E, Lucet JC, Timsit JF, Vanjak D, Paugam-Burtz C, Trouillet JL, et al. Acquisition of multidrug-resistant Pseudomonas aeruginosa in patients in intensive care units: role of antibiotics with antipseudomonal activity. Clin Infect Dis. 2004;38(5):670–7.
Park YS, Lee H, Chin BS, Han SH, Hong SG, Hong SK, et al. Acquisition of extensive drug-resistant Pseudomonas aeruginosa among hospitalized patients: risk factors and resistance mechanisms to carbapenems. J Hosp Infect. 2011;79(1):54–8.
Pena C, Gomez-Zorrilla S, Suarez C, Dominguez MA, Tubau F, Arch O, et al. Extensively drug-resistant Pseudomonas aeruginosa: risk of bloodstream infection in hospitalized patients. Eur J Clin Microbiol Infect Dis. 2012;31(10):2791–7.
Samonis G, Vardakas KZ, Kofteridis DP, Dimopoulou D, Andrianaki AM, Chatzinikolaou I, et al. Characteristics, risk factors and outcomes of adult cancer patients with extensively drug-resistant Pseudomonas aeruginosa infections. Infection. 2014;42(4):721–8.
Tumbarello M, Repetto E, Trecarichi EM, Bernardini C, De PG, Parisini A, et al. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: risk factors and mortality. Epidemiol Infect. 2011;139(11):1740–9.
Tuncer EG, Sonmezer MC, Tulek N, Erdinc SF, Bulut C, Berkem R, et al. Evaluation of risk factors for nosocomial multidrugresistant Pseudomonas aeruginosa infections. Clin Microbiol Infect. 2012;18:517. Conference: 22nd European Congress of Clinical Microbiology and Infectious Diseases London United Kingdom Conference Start: 20120331 Conference End: 20120403 Conference Publication: (var pagings).
Ustun C, Hosoglu S, Geyik MF. Risk factors for multi-drug-resistant Pseudomonas aeruginosa infections in a University Hospital - a case control study. Konuralp Tip Dergisi. 2016;8(2):2016.
Willmann M, Klimek AM, Vogel W, Liese J, Marschal M, Autenrieth IB, et al. Clinical and treatment-related risk factors for nosocomial colonisation with extensively drug-resistant Pseudomonas aeruginosa in a haematological patient population: a matched case control study. BMC Infect Dis. 2014;14:650.
Pena C, Suarez C, Tubau F, Dominguez A, Sora M, Pujol M, et al. Carbapenem-resistant Pseudomonas aeruginosa: factors influencing multidrug-resistant acquisition in non-critically ill patients. Eur J Clin Microbiol Infect Dis. 2009;28(5):519–22.
Tuon FF, Gortz LW, Rocha JL. Risk factors for pan-resistant Pseudomonas aeruginosa bacteremia and the adequacy of antibiotic therapy. Braz J Infect Dis. 2012;16(4):351–6.
Akhabue E, Synnestvedt M, Weiner MG, Bilker WB, Lautenbach E. Cefepime-Resistant Pseudomonas aeruginosa. Emerg Infect Dis. 2011;17(6):1037–43.
DalBen MF, Basso M, Garcia CP, Costa SF, Toscano CM, Jarvis WR, et al. Colonization pressure as a risk factor for colonization by multiresistant Acinetobacter spp and carbapenem-resistant Pseudomonas aeruginosa in an intensive care unit.[Erratum appears in Clinics (Sao Paulo). 2013 Dec;68(12):1559]. Clinics (Sao Paulo, Brazil). 2013;68(8):1128–33.
Djordjevic Z, Folic M, Ruzic ZD, Ilic G, Jankovic S. Risk factors for carbapenem-resistant Pseudomonas aeruginosa infection in a tertiary care hospital in Serbia. J Infect Dev Ctries. 2013;7(9):686–90.
Eagye KJ, Kuti JL, Nicolau DP. Risk factors and outcomes associated with isolation of meropenem high-level-resistant Pseudomonas aeruginosa. Infect Control Hosp Epidemiol. 2009;30(8):746–52.
Fortaleza CM, Freire MP, Filho DC, de Carvalho RM. Risk factors for recovery of imipenem- or ceftazidime-resistant pseudomonas aeruginosa among patients admitted to a teaching hospital in Brazil. Infect Control Hosp Epidemiol. 2006;27(9):901–6.
Furtado GH, Bergamasco MD, Menezes FG, Marques D, Silva A, Perdiz LB, et al. Imipenem-resistant Pseudomonas aeruginosa infection at a medical-surgical intensive care unit: risk factors and mortality. J Crit Care. 2009;24(4):625–14.
Furtado GH, Gales AC, Perdiz LB, Santos AE, Wey SB, Medeiros EA. Risk factors for hospital-acquired pneumonia caused by imipenem-resistant Pseudomonas aeruginosa in an intensive care unit. Anaesth Intensive Care. 2010;38(6):994–1001.
Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am J Med. 2006;119(6):526–5.
Harris AD, Perencevich E, Roghmann MC, Morris G, Kaye KS, Johnson JA. Risk factors for piperacillin-tazobactam-resistant Pseudomonas aeruginosa among hospitalized patients. Antimicrob Agents Chemother. 2002;46(3):854–8.
Harris AD, Smith D, Johnson JA, Bradham DD, Roghmann MC. Risk factors for imipenem-resistant Pseudomonas aeruginosa among hospitalized patients. Clin Infect Dis. 2002;34(3):340–5.
Harris AD, Johnson JK, Thom KA, Morgan DJ, McGregor JC, Ajao AO, et al. Risk factors for development of intestinal colonization with imipenem-resistant Pseudomonas aeruginosa in the intensive care unit setting. Infect Control Hosp Epidemiol. 2011;32(7):719–22.
Hsu DI, Okamoto MP, Murthy R, Wong-Beringer A. Fluoroquinolone-resistant Pseudomonas aeruginosa: risk factors for acquisition and impact on outcomes. J Antimicrob Chemother. 2005;55(4):535–41.
Khayr W, Rheault W, Waiters L, Walters A. Epidemiology of ciprofloxacin-resistant Pseudomonas aeruginosa in a veterans affairs hospital. Am J Ther. 2000;7(5):309–12.
Kohlenberg A, Weitzel-Kage D, van der Linden P, Sohr D, Vogeler S, Kola A, et al. Outbreak of carbapenem-resistant Pseudomonas aeruginosa infection in a surgical intensive care unit. J Hosp Infect. 2010;74(4):350–7.
Lautenbach E, Synnestvedt M, Weiner MG, Bilker WB, Vo L, Schein J, et al. Imipenem resistance in Pseudomonas aeruginosa: emergence, epidemiology, and impact on clinical and economic outcomes. Infect Control Hosp Epidemiol. 2010;31(1):47–53.
Lee CH, Su TY, Ye JJ, Hsu PC, Kuo AJ, Chia JH, et al. Risk factors and clinical significance of bacteremia caused by Pseudomonas aeruginosa resistant only to carbapenems. J Microbiol Immunol Infect. 2017;50(5):677–83.
Lin KY, Lauderdale TL, Wang JT, Chang SC. Carbapenem-resistant Pseudomonas aeruginosa in Taiwan: prevalence, risk factors, and impact on outcome of infections. J Microbiol Immunol Infect. 2016;49(1):52–9.
Luyt CE, Aubry A, Lu Q, Micaelo M, Brechot N, Brossier F, et al. Imipenem, meropenem, or doripenem to treat patients with Pseudomonas aeruginosa ventilator-associated pneumonia. Antimicrob Agents Chemother. 2014;58(3):1372–80.
Onguru P, Erbay A, Bodur H, Baran G, Akinci E, Balaban N, et al. Imipenem-resistant Pseudomonas aeruginosa: risk factors for nosocomial infections. J Korean Med Sci. 2008;23(6):982–7.
Pena C, Guzman A, Suarez C, Dominguez MA, Tubau F, Pujol M, et al. Effects of carbapenem exposure on the risk for digestive tract carriage of intensive care unit-endemic carbapenem-resistant Pseudomonas aeruginosa strains in critically ill patients. Antimicrob Agents Chemother. 2007;51(6):1967–71.
Pereira GH, Levin AS, Oliveira HB, Moretti ML. Controlling the clonal spread of Pseudomonas aeruginosa infection. Infect Control Hosp Epidemiol. 2008;29(6):549–52.
Tam VH, Chang KT, LaRocco MT, Schilling AN, McCauley SK, Poole K, et al. Prevalence, mechanisms, and risk factors of carbapenem resistance in bloodstream isolates of Pseudomonas aeruginosa. Diagn Microbiol Infect Dis. 2007;58(3):309–14.
Trouillet JL, Vuagnat A, Combes A, Kassis N, Chastre J, Gibert C. Pseudomonas aeruginosa ventilator-associated pneumonia: comparison of episodes due to piperacillin-resistant versus piperacillin-susceptible organisms. Clin Infect Dis. 2002;34(8):1047–54.
Zavascki AP, Cruz RP, Goldani LZ. Risk factors for imipenem-resistant Pseudomonas aeruginosa: a comparative analysis of two case-control studies in hospitalized patients. J Hosp Infect. 2005;59(2):96–101.
Zhang JF, Chen BL, Xin XY, Zhao HB, Wang HY, Song H, et al. Carbapenem resistance mechanism and risk factors of Pseudomonas aeruginosa clinical isolates from a University Hospital in Xi'an, China. Microb Drug Resist. 2009;15(1):41–5.
Nicasio AM, Kuti JL, Nicolau DP. The current state of multidrug-resistant gram-negative bacilli in North America. Pharmacotherapy. 2008;28(2):235–49.
Buhl M, Peter S, Willmann M. Prevalence and risk factors associated with colonization and infection of extensively drug-resistant Pseudomonas aeruginosa: a systematic review. Expert Rev Anti-Infect Ther. 2015;13(9):1159–70.
Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Funding
Funding for this review was provided to Tufts Medical Center by the Merck & Co., Inc., Kenilworth, NJ, USA. The funder played no role in the study search and selection, data synthesis and analysis.
Availability of data and materials
Data supporting the conclusions of this article is available in the Supplementary Material.
Author information
Authors and Affiliations
Contributions
SM designed research; GR, EEA, and JC conducted research; GR and EEA analyzed data; and GR, EEA, and JC wrote the paper; SM and LP edited the paper; GR, EEA, and JC had primary responsibility for final content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable
Consent for publication
Not Applicable
Competing interests
S. Merchant and L. Puzniak are employees of Merck & Co., Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Search Strategy. Table S2. Patient–related Multivariate Risk Factors of Acquistion of MDR P. aeruginosa. Table S3. Antibiotic Treatment–related Multivariate Risk Factors of Acquistion of MDR P. aeruginosa. Table S4. Other Treatment–related Multivariate Risk Factors of Acquistion of MDR and XDR P. aeruginosa. Table S5. Hospital–related Multivariate Risk Factors of Acquistion of MDR and XDR P. aeruginosa. Table S6. Patient–related Multivariate Risk Factors of Acquistion of Carbapenem-resistant P. aeruginosa. Table S7. Antibiotic Treatment–related Multivariate Risk Factors of Acquistion of Carbapenem-resistant P. aeruginosa. Table S8. Other Treatment–related Multivariate Risk Factors of Acquistion of Carbapenem-resistant P. aeruginosa. Table S9. Hospital–related Multivariate Risk Factors of Acquistion of Carbapenem-resistant P. aeruginosa. Table S10. Multivariate Risk Factors of Acquistion of Resistant P. aeruginosa. Figure S1. Meta-analysis of Risk Factors for Carbapenem versus Susceptible P. aeruginosa Acquisition. Figure S2. Meta-analysis of Prior Use of Carbapenem as a Risk Factor for Carbapenem versus Susceptible P. aeruginosa Acquisition. Figure S3. Meta-analysis of Prior Use of Fluoroquinolones as a Risk Factor for Quinolone-resistant versus Susceptible P. aeruginosa Acquisition. (DOCX 182 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Raman, G., Avendano, E., Chan, J. et al. Risk factors for hospitalized patients with resistant or multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 7, 79 (2018). https://doi.org/10.1186/s13756-018-0370-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-018-0370-9