Abstract
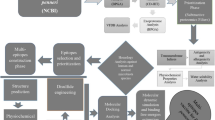
Infectious serositis of ducks, caused by Riemerella anatipestifer, is one of the main infectious diseases that harm commercial ducks. Whole-strain-based vaccines with no or few cross-protection were observed between different serotypes of R. anatipestifer, and so far, control of infection is hampered by a lack of effective vaccines, especially subunit vaccines with cross-protection. Since the concept of reverse vaccinology was introduced, it has been widely used to screen for protective antigens in important pathogens. In this study, pan-genome binding reverse vaccinology, an emerging approach to vaccine candidate screening, was used to screen for cross-protective antigens against R. anatipestifer. Thirty proteins were identified from the core-genome as potential cross-protective antigens. Three of these proteins were recombinantly expressed, and their immunoreactivity with five antisera (anti-serotypes 1, 2, 6, 10, and 11) was demonstrated by Western blotting. Our study established a method for high-throughput screening of cross-protective antigens against R. anatipestifer in silico, which will lay the foundation for the development of a cross-protective subunit vaccine controlling R. anatipestifer infection.
Key points
• Pan-genome binding reverse vaccine approach was first established in R. anatipestifer to screen for subunit vaccine candidates.
• Thirty potential cross-protective antigens against R. anatipestifer were identified by this method.
• The reliability of the method was verified preliminarily by the results of Western blotting of three of these potential antigens.







Similar content being viewed by others
Data availability
The genome datasets generated during the current study are available in the NCBI database, [https://www.ncbi.nlm.nih.gov/genome/browse/#!/prokaryotes/2536/]. All data generated or analyzed during this study are included in this manuscript.
References
Cha S-Y, Seo H-S, Wei B, Kang M, Roh J-H, Yoon R-H, Kim J-H, Jang H-K (2015) Surveillance and characterization of Riemerella anatipestifer from wild birds in South Korea. J Wildl Dis 51:341–347. https://doi.org/10.7589/2014-05-128
Chang F-F, Chen C-C, Wang S-H, Chen C-L (2019) Epidemiology and antibiogram of isolated from waterfowl slaughterhouses in Taiwan. J Vet Res 63:79–86. https://doi.org/10.2478/jvetres-2019-0003
Chen Z, Wang X, Ren X, Han W, Malhi KK, Ding C, Yu S (2019) Riemerella anatipestifer GldM is required for bacterial gliding motility, protein secretion, and virulence. Vet Res 50:43. https://doi.org/10.1186/s13567-019-0660-0
Confer AW, Ayalew S (2013) The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163:207–222. https://doi.org/10.1016/j.vetmic.2012.08.019
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8:4. https://doi.org/10.1186/1471-2105-8-4
Gao Q, Lu S, Wang M, Jia R, Chen S, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Mao S, Ou X, Sun D, Tian B, Cheng A (2021) Putative Riemerella anatipestifer outer membrane protein H affects virulence. Front Microbiol 12
Grandi G (2010) Bacterial surface proteins and vaccines F1000 Biol Rep 2 36 https://doi.org/10.3410/B2-36
Guo Y, Hu D, Guo J, Wang T, Xiao Y, Wang X, Li S, Liu M, Li Z, Bi D, Zhou Z (2017) Riemerella anatipestifer type IX secretion system is required for virulence and gelatinase secretion. Front Microbiol 8
Gurevich A, Saveliev V, Vyahhi N, Tesler G (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. https://doi.org/10.1093/bioinformatics/btt086
Gyuris É, Wehmann E, Czeibert K, Magyar T (2017) Antimicrobial susceptibility of Riemerella anatipestifer strains isolated from geese and ducks in Hungary. Acta Vet Hung 65:153–165. https://doi.org/10.1556/004.2017.016
Hallgren J, Tsirigos KD, Pedersen MD, Armenteros JJA, Marcatili P, Nielsen H, Krogh A, Winther O (2022) Deep TMHMM predicts alpha and beta transmembrane proteins using deep neural networks 04 08 487609
Han X, Hu Q, Ding S, Chen W, Ding C, He L, Wang X, Ding J, Yu S (2012) Identification and immunological characteristics of chaperonin GroEL in Riemerella anatipestifer. Appl Microbiol Biotechnol 93:1197–1205. https://doi.org/10.1007/s00253-011-3635-2
Hu Q, Han X, Zhou X, Ding C, Zhu Y, Yu S (2011) OmpA is a virulence factor of Riemerella anatipestifer. Vet Microbiol 150:278–283. https://doi.org/10.1016/j.vetmic.2011.01.022
Hu Q, Ding C, Tu J, Wang X, Han X, Duan Y, Yu S (2012) Immunoproteomics analysis of whole cell bacterial proteins of Riemerella anatipestifer. Vet Microbiol 157:428–438. https://doi.org/10.1016/j.vetmic.2012.01.009
Hu D, Guo Y, Guo J, Wang Y, Pan Z, Xiao Y, Wang X, Hu S, Liu M, Li Z, Bi D, Zhou Z (2019) Deletion of the Riemerella anatipestifer type IX secretion system gene sprA results in differential expression of outer membrane proteins and virulence. Avian Pathol 48:191–203. https://doi.org/10.1080/03079457.2019.1566594
Huang B, Subramaniam S, Frey J, Loh H, Tan H-M, Fernandez CJ, Kwang J, Chua K-L (2002) Vaccination of ducks with recombinant outer membrane protein (OmpA) and a 41 kDa partial protein (P45N′) of Riemerella anatipestifer. Vet Microbiol 84:219–230. https://doi.org/10.1016/S0378-1135(01)00456-4
Jadhav A, Shanmugham B, Rajendiran A, Pan A (2014) Unraveling novel broad-spectrum antibacterial targets in food and waterborne pathogens using comparative genomics and protein interaction network analysis. Infect Genet Evol 27:300–308. https://doi.org/10.1016/j.meegid.2014.08.007
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. https://doi.org/10.1038/s41467-018-07641-9
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Kang M, Seo H-S, Soh S-H, Jang H-K (2018) Immunogenicity and safety of a live Riemerella anatipestifer vaccine and the contribution of IgA to protective efficacy in Pekin ducks. Vet Microbiol 222:132–138. https://doi.org/10.1016/j.vetmic.2018.07.010
Koebnik R, Locher KP, Van Gelder P (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37:239–253. https://doi.org/10.1046/j.1365-2958.2000.01983.x
Mavromatis K, Lu M, Misra M, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng J-F, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Pagani I, Ivanova N, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Jeffries CD, Detter JC, Brambilla E-M, Rohde M, Göker M, Gronow S, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk H-P, Kyrpides NC (2011) Complete genome sequence of Riemerella anatipestifer type strain (ATCC 11845T). Stand Genomic Sci 4:144–153. https://doi.org/10.4056/sigs.1553862
Mora M, Veggi D, Santini L, Pizza M, Rappuoli R (2003) Reverse vaccinology. Drug Discov Today 8:459–464. https://doi.org/10.1016/S1359-6446(03)02689-8
Naz A, Awan FM, Obaid A, Muhammad SA, Paracha RZ, Ahmad J, Ali A (2015) Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect Genet Evol 32:280–291. https://doi.org/10.1016/j.meegid.2015.03.027
Nhung NT, Chansiripornchai N, Carrique-Mas JJ (2017) Antimicrobial resistance in bacterial poultry pathogens a review. Front Vet Sci 4
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. https://doi.org/10.1093/bioinformatics/btv421
Pathanasophon P, Sawada T, Tanticharoenyos T (1995) New serotypes of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 24:195–199. https://doi.org/10.1080/03079459508419059
Pathanasophon P, Sawada T, Pramoolsinsap T, Tanticharoenyos T (1996) Immunogenicity of Riemerella anatipestifer broth culture bacterin and cell-free culture filtrate in ducks. Avian Pathol 25:705–719. https://doi.org/10.1080/03079459608419176
Pathanasophon P, Phuektes P, Tanticharoenyos T, Narongsak W, Sawada T (2002) A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol 31:267–270. https://doi.org/10.1080/03079450220136576
Sandhu T (1979) Immunization of White Pekin ducklings against Pasteurella anatipestifer infection. Avian Dis 23:662–669
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Segers P, Mannheim W, Vancanneyt M, De Brandt K, Hinz K-H, Kersters K, Vandamme PY (1993) Riemerella anatipestifer gen. nov., comb. nov., the Causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int J Syst Evol Microbiol 43:768–776. https://doi.org/10.1099/00207713-43-4-768
Smith SGJ, Mahon V, Lambert MA, Fagan RP (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 273:1–11. https://doi.org/10.1111/j.1574-6968.2007.00778.x
Subramaniam S, Huang B, Loh H, Kwang J, Tan H-M, Chua K-L, Frey J (2000) Characterization of a predominant immunogenic outer membrane protein of Riemerella anatipestifer. Clin Diagn Lab Immunol 7:168–174. https://doi.org/10.1128/CDLI.7.2.168-174.2000
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol 38:3022. https://doi.org/10.1093/molbev/msab120
Tettelin H (2009) The bacterial pan-genome and reverse vaccinology. Microb Pathog 6:35–47. https://doi.org/10.1159/000235761
Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, DeBoy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJB, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci 102:13950–13955. https://doi.org/10.1073/pnas.0506758102
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Žídek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S (2021) AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. https://doi.org/10.1093/nar/gkab1061
Veith PD, Glew MD, Gorasia DG, Reynolds EC (2017) Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol 106:35–53. https://doi.org/10.1111/mmi.13752
Wang X, Liu W, Zhu D, Yang L, Liu M, Yin S, Wang M, Jia R, Chen S, Sun K, Cheng A, Chen X (2014) Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:1–10. https://doi.org/10.1186/1471-2164-15-479
Wick RR, Judd LM, Gorrie CL, Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol 13:e1005595. https://doi.org/10.1371/journal.pcbi.1005595
Xu X, Xu Y, Miao S, Jiang P, Cui J, Gong Y, Tan P, Du X, Islam N, Hu Q (2020) Evaluation of the protective immunity of Riemerella anatipestifer OmpA. Appl Microbiol Biotechnol 104:1273–1281. https://doi.org/10.1007/s00253-019-10294-3
Yang S, Dong W, Li G, Zhao Z, Song M, Huang Z, Fu J, Jia F, Lin S (2019) A recombinant vaccine of Riemerella anatipestifer OmpA fused with duck IgY Fc and Schisandra chinensis polysaccharide adjuvant enhance protective immune response. Microb Pathog 136:103707. https://doi.org/10.1016/j.micpath.2019.103707
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. https://doi.org/10.1093/bioinformatics/btq249
Zagursky RJ, Olmsted SB, Russell DP, Wooters JL (2003) Bioinformatics: how it is being used to identify bacterial vaccine candidates. Expert Rev Vaccines 2:417–436. https://doi.org/10.1586/14760584.2.3.417
Zhai Z, Li X, Xiao X, Yu J, Chen M, Yu Y, Wu G, Li Y, Ye L, Yao H, Lu C, Zhang W (2013) Immunoproteomics selection of cross-protective vaccine candidates from Riemerella anatipestifer serotypes 1 and 2. Vet Microbiol 162:850–857. https://doi.org/10.1016/j.vetmic.2012.11.002
Zhong CY, Cheng AC, Wang MS, Zhu DK, Luo QH, Zhong CD, Li L, Duan Z (2009) Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis 53:601–607. https://doi.org/10.1637/8552-120408-ResNote.1
Zhou Z, Peng X, Xiao Y, Wang X, Guo Z, Zhu L, Liu M, Jin H, Bi D, Li Z, Sun M (2011) Genome sequence of poultry pathogen Riemerella anatipestifer strain RA-YM. J Bacteriol 193:1284–1285. https://doi.org/10.1128/JB.01445-10
Zhu D, Yang Z, Xu J, Wang M, Jia R, Chen S, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Chen X, Cheng A (2020) Pan-genome analysis of Riemerella anatipestifer reveals its genomic diversity and acquired antibiotic resistance associated with genomic islands. Funct Integr Genomics 20:307–320. https://doi.org/10.1007/s10142-019-00715-x
Funding
This work was supported by the Project of Sanya Yazhou Bay Science and Technology City, Grant No. SCKJ-JYRC-2022–80; The Guidance Foundation, the Sanya Institute of Nanjing Agricultural University (NAUSY-MS12); Natural Science Foundation of Shandong Province (ZR2020MC175); and Major Scientific and Technological Innovation Projects in Shandong Province (2019JZZY010719).
Author information
Authors and Affiliations
Contributions
X.K. Z. conceived and designed research. X.K. Z. and S.X. X. conducted experiments. Z.H. W. and L. D. contributed reagents and analytical tools. X.K. Z. analyzed data. X.K. Z. and X.Y. T. wrote and revised the manuscript. Y.Q. L., Y.B. L., and W. Z. applied for funding and supervised research. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All experimentation was conducted following permission from the Ministry of Science and Technology of Jiangsu Province, China.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zheng, X., Xu, S., Wang, Z. et al. Sifting through the core-genome to identify putative cross-protective antigens against Riemerella anatipestifer. Appl Microbiol Biotechnol 107, 3085–3098 (2023). https://doi.org/10.1007/s00253-023-12479-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12479-3