Abstract
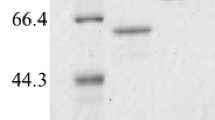
In this study, a novel high-activity amylosucrase from Salinispirillum sp. LH10-3-1 (SaAS) was identified and characterized. The recombinant enzyme was determined as a monomer with a molecular mass of 75 kDa. SaAS protein exhibited the maximum total and polymerization activities at pH 9.0 and maximum hydrolysis activity at pH 8.0. The optimum temperature for total, polymerization, and hydrolysis activities were 40, 40, and 45 °C, respectively. Under the optimal pH and temperature, SaAS had a specific activity of 108.2 U/mg. SaAS also showed excellent salt tolerance and could retain 77.4% of its original total activity at 4.0 M NaCl. The addition of Mg2+, Ba2+, and Ca2+ enhanced the total activity of SaAS. When the conversion of 0.1 M and 1.0 M sucrose was catalyzed at pH 9.0 and 40 °C for 24 h, the ratios of hydrolysis, polymerization, and isomerization reactions were 11.9:77.4:10.7 and 15.3:53.5:31.2, respectively. The α-arbutin yield of 60.3% was achieved from 20 mM sucrose and 5 mM hydroquinone catalyzed by SaAS.
Key points
• A novel amylosucrase from Salinispirillum sp. LH10-3-1 (SaAS) was characterized.
• SaAS has the highest specific enzyme activity among all known amylosucrase.
• SaAS has hydrolysis, polymerization, isomerization, and glucosyltransferase activities.
Graphical Abstract








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this manuscript.
References
Agarwal N, Narnoliya LK, Singh SP (2019) Characterization of a novel amylosucrase gene from the metagenome of a thermal aquatic habitat, and its use in turanose production from sucrose biomass. Enzyme Microb Technol 131:109372
But SY, Khmelenina VN, Reshetnikov AS, Mustakhimov II, Kalyuzhnaya MG, Trotsenko YA (2015) Sucrose metabolism in halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Arch Microbiol 197:471–480
Cambon E, Barbe S, Pizzut-Serin S, Remaud-Simeon M, André I (2014) Essential role of amino acid position 226 in oligosaccharide elongation by amylosucrase from Neisseria polysaccharea. Biotechnol Bioeng 111:1719–1728
Chin YW, Jang SW, Shin HS, Kim TW, Kim SK, Park CS, Seo DH (2020) Heterologous expression of Deinococcus geothermalis amylosucrase in Corynebacterium glutamicum for luteolin glucoside production. Enzyme Microb Technol 135:109505
Choi SW, Lee JA, Yoo SH (2019) Sucrose-based biosynthetic process for chain-lengthdefined α-glucan and functional sweetener by Bifidobacterium amylosucrase. Carbohydr Polym 205:581–588
De Montalk GP, Remaud-Simeon M, Willemot RM, Sarabal P, Planchot V, Monsan P (2000) Amylosucrase from Neisseria polysaccharea: novel catalytic properties. FEBS Lett. 471:219–223
Emond S, Mondeil S, Jaziri K, André I, Monsan P, Remaud-Siméon M, Potocki-Véronèse G (2008) Cloning, purification and characterization of a thermostable amylosurase from Deinococcus geothermalis. FEMS Microbiol Lett 285:25–32
Guérin F, Barbe S, Pizzut-Serin S, Potocki-Véronèse G, Guieysse D, Guillet V, Monsan P, Mourey L, Remaud-Siméon M, André I, Tranier S (2012) Structural investigation of the thermostability and product specificity of amylosucrase from the bacterium Deinococcus geothermalis. J Biol Chem 287:6642–6654
Ha SJ, Seo DH, Jung JH, Cha J, Kim TJ, Kim YW, Park CS (2009) Molecular cloning and functional expression of a new amylosucrase from Alteromonas macleodii. Biosci Biotechnol Biochem 73:1505–1512
Hehre EJ, Hamilton DM (1946) Bacterial synthesis of an amylopectin-like polysaccharide from sucrose. J Biol Chem 166:777–778
Jeong JW, Seo DH, Jung JH, Park JH, Baek NI, Kim MJ, Park CS (2014) Biosynthesis of glucosyl glycerol, a compatible solute, using intermolecular transglycosylation activity of amylosucrase from Methylobacillus flagellatus KT. Appl Biochem Biotechnol 173:904–917
Jung JH, Seo DH, Ha SJ, Song MC, Cha J, Yoo SH, Kim TJ, Baek NI, Baik MY, Park CS (2009) Enzymatic synthesis of salicin glycosides through transglycosylation catalyzed by amylosucrases from Deinococcus geothermalis and Neisseria polysaccharea. Carbohydr Res 344:1612–1619
Kim MD, Seo DH, Jung JH, Jung DH, Joe MH, Lim S, Lee JH, Park CS (2014) Molecular cloning and expression of amylosucrase from highly radiation-resistant Deinococcus radiopugnans. Food Sci Biotechnol 23:2007–2012
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Li J, Li Z, Cao M, Liu J (2021) Expression and characterization of catechol 1,2-dioxygenase from Oceanimonas marisflavi 102-Na3. Protein Expr Purif 188:105964
Miao M, Ma YJ, Jiang B, Huang C, Li XH, Cui SW, Zhang T (2014) Structural investigation of a neutral extracellular glucan from Lactobacillus reuteri SK24.003. Carbohydr Polym 106:384–392
Park KH (2008) Carbohydrate-active enzyme structure, function and application. CRC press, Boca Raton
Park MO, Chandrasekaran M, Yoo SH (2018) Expression, purification, and characterization of a novel amylosucrase from Neisseria subflava. Int J Biol Macromol 109:160–166
Perez-Cenci M, Salerno GL (2014) Functional characterization of Synechococcus amylosucrase and fructokinase encoding genes discovers two novel actors on the stage of cyanobacterial sucrose metabolism. Plant Sci 224:95–102
Pizzut-Serin S, Potocki-Véroènse G, van der Veen BA, Albenne C, Monsan P, Remaud-Simeon M (2005) Characterisation of a novel amylosucrase from Deinococcus radiodurans. FEBS Lett 579:1405–1410
Rha CS, Choi JM, Jung YS, Kim ER, Ko MJ, Seo DH, Kim DO, Park CS (2019) High-efficiency enzymatic production of α-isoquercitrin glucosides by amylosucrase from Deinococcus geothermalis. Enzyme Microb Technol 120:84–90
Rha CS, Kim HG, Baek NI, Kim DO, Park CS (2020) Amylosucrase from Deinococcus geothermalis can be modulated under different reaction conditions to produce novel quercetin 4′-O-α-d-isomaltoside. Enzyme Microb Technol 141:109648
Schneider J, Fricke C, Overwin H, Hofer B (2011) High level expression of a recombinant amylosucrase gene and selected properties of the enzyme. Appl Microbiol Biotechnol 89:1821–1829
Sedmak JJ, Grossberg SE (1977) A rapid, sensitive and vertile assay for protein using Coomassia Brilliant Blue G250. Anal Biochem 79:544–552
Seo DH, Jung JH, Choi HC, Cho HK, Kim HH, Ha SJ, Yoo SH, Cha J, Park CS (2012a) Functional expression of amylosucrase, a glucan-synthesizing enzyme, from Arthrobacter chlorophenolicus A6. J Microbiol Biotechnol 22:1253–1257
Seo DH, Jung JH, Ha SJ, Cho HK, Jung DH, Kim TJ, Baek NI, Yoo SH, Park CS (2012b) High-yield enzymatic bioconversion of hydroquinone to α-arbutin, a powerful skin lightening agent, by amylosucrase. Appl Microbiol Biotechnol 94:1189–1197
Seo DH, Jung JH, Jung DH, Park S, Yoo SH, Kim YR, Park CS (2016) An unusual chimeric amylosucrase generated by domain-swapping mutagenesis. Enzyme Microb Technol 86:7–16
Seo DH, Yoo SH, Choi SJ, Kim YR, Park CS (2020) Versatile biotechnological application of amylosucrase, a novel glucosyltransferase. Food Sci Biotechnol 29:1–16
Skov LK, Mirza O, Henriksen A, De Montalk GP, Remaud-Simeon M, Sarcabal P, Willemot RM, Monsan P, Gajhede M (2000) Crystallization and preliminary X-ray studies of recombinant amylosucrase from Neisseria polysaccharea. Acta Cryst 56:203–205
Skov LK, Mirza O, Henriksen A, De Montalk GP, Remaud-Simeon M, Sarcabal P, Willemot RM, Monsan P, Gajhede M (2001) Amylosucrase: aglucan-synthesizing enzyme from the α-amylase family. J Biol Chem 276:25273–25278
Skov LK, Mirza O, Sprogøe D, Dar I, Remaud-Simeon M, Albenne C, Monsan P, Gajhede M (2002) Oligosaccharide and sucrose complexes ofamylosucrase: structural implications for the polymerase activity. J Biol Chem 277:47741–47747
Skov LK, Pizzut-Serin S, Remaud-Simeon M, Ernst HA, Gajhede M, Mirza O (2013) The structure of amylosucrase from Deinococcus radiodurans has an unusual openactive-site topology. Acta Crystallogr Sect F Struct Biol Cryst Commun 69:973–978
Tian Y, Xu W, Zhang W, Zhang T, Guang C, Mu W (2018) Amylosucrase as a transglucosylation tool: from molecular features to bioengineering applications. Biotechnol Adv 36:1540–1552
Tian Y, Xu W, Guang C, Zhang W, Mu W (2019) Thermostable amylosucrase from Calidithermus timidus DSM 17022: insight into its characteristics and tetrameric conformation. J Agric Food Chem 67:9868–9876
Wang R, Kim JH, Kim BS, Park CS, Yoo SH (2011) Preparation and characterization of non-covalently immobilized amylosucrase using a pH-dependent autoprecipitating carrier. Bioresource Technol 102:6370–6374
Wang Y, Xu W, Bai Y, Zhang T, Jiang B, Mu W (2017) Identification of an α-(1,4)-glucan-synthesizing amylosucrase from Cellulomonas carboniz T26. J Agric Food Chem 65:2110–2119
Xi L, Tan W, Li J, Qu J, Liu J (2021) Cloning and characterization of a novel thermostable amidase, Xam, from Xinfangfanggia sp. DLY26. Biotechnol Lett 43:1395–1402
Yang C, Fan W, Zhang R, Shi J, Knežević-Jugović Z, Zhang B (2019) Study on transglucosylation properties of amylosucrase from Xanthomonas campestris pv. campestris and its application in the production of α-arbutin. Catalysts 9: 5
Yu S, Wang Y, Tian Y, Xu W, Bai Y, Zhang T, Mu W (2018) Highly efficient biosynthesis of α-arbutin from hydroquinone by an amylosucrase from Cellulomonas carboniz. Food Biochem 68:93–99
Zhang T, Li Y, Zhou Y, Zhang L (2016) Cloning and expression analysis of a homologous expansin gene EXP2 in Picea wilsonii. J For Res 27:247–255
Zhu X, Tian Y, Xu W, Bai Y, Zhang T, Mu W (2018) Biochemical characterization of a highly thermostable amylosucrase from Truepera radiovictrix DSM 17093. Int J Biol Macromol 116:744–752
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 21473256 and 21776310), the Key Research and Development Project of Shandong Province (No. 2019GSF107077), and the Key Technology of Independent Innovation of Qingdao West Coast New Economic District (2020-6).
Author information
Authors and Affiliations
Contributions
JL designed the study, JL, ZL, HG, and MM conducted the experiments and analyzed data. SL, HY, and HZ assisted the experiments and analyzed data. JL and JL wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Li, Z., Gong, H. et al. Identification and characterization of a novel high-activity amylosucrase from Salinispirillum sp. LH10-3-1. Appl Microbiol Biotechnol 107, 1725–1736 (2023). https://doi.org/10.1007/s00253-023-12430-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-023-12430-6