Abstract
Herein, two genes (LBA0625 and LBA1719) encoding UGPases (UDP-glucose pyrophosphorylase) in Lactobacillus acidophilus (L. acidophilus) were successfully transformed into Escherichia coli BL21 (DE3) to construct recombinant overexpressing strains (E-0625, E-1719) to investigate the biological characteristics of UGPase-0625 and UGPase-1719. The active sites, polysaccharide yield, and anti-freeze-drying stress of L. acidophilus ATCC4356 were also detected. UGPase-0625 and UGPase-1719 belong to the nucleotidyltransferase of stable hydrophilic proteins; contain 300 and 294 amino acids, respectively; and have 20 conserved active sites by prediction. Αlpha-helixes and random coils were the main secondary structures, which constituted the main skeleton of UGPases. The optimal mixture for the high catalytic activity of the two UGPases included 0.5 mM UDP-Glu (uridine diphosphate glucose) and Mg2+ at 37 °C, pH 10.0. By comparing the UGPase activities of the mutant strains with the original recombinant strains, A10, L130, and L263 were determined as the active sites of UGPase-0625 (P < 0.01) and A11, L130, and L263 were determined as the active sites of UGPase-1719 (P < 0.01). In addition, UGPase overexpression could increase the production of polysaccharides and the survival rates of recombinant bacteria after freeze-drying. This is the first study to determine the enzymatic properties, active sites, and structural simulation of UGPases from L. acidophilus, providing in-depth understanding of the biological characteristics of UGPases in lactic acid bacteria.
Key points
• We detected the biological characteristics of UGPases encoded by LBA0625 and LBA1719.
• We identified UGPase-0625 and UGPase-1719 active sites.
• UGPase overexpression elevates polysaccharide levels and post-freeze-drying survival.
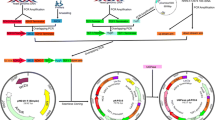
Graphical abstract






Similar content being viewed by others
Availability of data and material
All data related to this study has been included in the manuscript.
References
Bais VS, Aggarwal P, Bharadwaj P, Prakash B (2020) Classification, characterization and structural analysis of sugar nucleotidylyltransferase family of enzymes. Biochem Biophys Res Commun 525(3):780–785. https://doi.org/10.1016/j.bbrc.2020.02.148
Balan D, Tokas J, Singal HR (2018) UDP-glucose pyrophosphorylase: isolation, purification and characterization from developing thermotolerant wheat (Triticum aestivum) grains. Protein Expr Purif 148:68–77. https://doi.org/10.1016/j.pep.2018.04.007
Banerji A, Ghosh I (2009) A new computational model to study mass inhomogeneity and hydrophobicity inhomogeneity in proteins. Eur Biophys J 38(5):577–587. https://doi.org/10.1007/s00249-009-0409-1
Baralle FE, Giudice J (2017) Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18(7):437–451. https://doi.org/10.1038/nrm.2017.27
Bhatwa A, Wang W, Hassan YI, Abraham N, Zhou T (2021) Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Frontiers in Bioengineering and Biotechnology 9:630551. https://doi.org/10.3389/fbioe.2021.630551
Bradford M (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Chen J, Wang L, Liang H, Jin X, Tian M (2020) Overexpression of DoUGP enhanced biomass and stress tolerance by promoting polysaccharide accumulation in Dendrobium officinale. Frontiers in Plant Science 11. https://doi.org/10.3389/fpls.2020.533767
Chen H, Chen S, Li C, Shu G (2015) Response surface optimization of lyoprotectant for Lactobacillus bulgaricus during vacuum freeze-drying. Prep Biochem Biotechnol 45(5):463–475. https://doi.org/10.1080/10826068.2014.923451
Chen MJ, Tang HY, Chiang ML (2017) Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol 66:20. https://doi.org/10.1016/j.fm.2017.03.020
Chi S, Feng YJ, Tao L (2015) Molecular cloning, characterization, and comparison of UDP-glucose pyrophosphorylase from Gracilaria chouae and Saccharina japonica. J Appl Phycol 28(3):2051–2059. https://doi.org/10.1007/s10811-015-0738-7
Cotrim CA, Soares J, Bostjan K, Marcelo M, Permyakov EA (2018) Crystal structure and insights into the oligomeric state of UDP-glucose pyrophosphorylase from sugarcane. Plos One 13(3). https://doi.org/10.1371/journal.pone.0193667
Decker D, Meng M, Gornicka A, Hofer A, Wilczynska M, Kleczkowski LA (2012) Substrate kinetics and substrate effects on the quaternary structure of barley UDP-glucose pyrophosphorylase. Phytochemistry 79:39–45. https://doi.org/10.1016/j.phytochem.2012.04.002
Dong MY, Fan XW, Li YZ (2019) Cassava AGPase genes and their encoded proteins are different from those of other plants. Planta 250(5):1621–1635. https://doi.org/10.1007/s00425-019-03247-7
Geisler M, Wilczynska M, Karpinski S, Kleczkowski LA (2004) Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: homology-modeling analyses. Plant Mol Biol 56(5):783–794. https://doi.org/10.1007/s11103-004-4953-x
Hougland JL, Kravchuk AV, Herschlag D, Piccirilli JA (2005) Functional identification of catalytic metal ion binding sites within RNA. PLoS Biol 3(9):1536–1548. https://doi.org/10.1371/journal.pbio.0030277
Imai Y, Moralez A, Andag U, Clarke JB, Busby WH, Clemmons DR (2020) Substitutions for hydrophobic amino acids in the N-terminal domains of IGFBP-3 and -5 markedly reduce IGF-I binding and alter their biologic actions. J Biol Chem 275(24):18188–18194. https://doi.org/10.1074/jbc.M000070200
Kathuria SV, Chan YH, Nobrega RP, Özen A, Matthews CR (2016) Clusters of isoleucine, leucine, and valine side chains define cores of stability in high-energy states of globular proteins: sequence determinants of structure and stability. Protein Sci 25(3):662–675. https://doi.org/10.1002/pro.2860
Kim H, Choi J, Kim T, Lokanath NK, Ha SC, Suh SW, Hwang HY, Kim KK (2010) Structural basis for the reaction mechanism of UDP-glucose pyrophosphorylase. Mol Cells 29(4):397–405. https://doi.org/10.1007/s10059-010-0047-6
Kleczkowski LA, Decker D (2015) Sugar activation for production of nucleotide sugars as substrates for glycosyltransferases in plants. J Appl Glycosci 62(2):28–32. https://doi.org/10.5458/jag.jag.JAG-2015_003
Kleczkowski LA, Martz F, Wilczynska M (2005) Factors affecting oligomerization status of UDP-glucose pyrophosphorylase. Phytochemistry 66(24):2815–2821. https://doi.org/10.1016/j.phytochem.2005.09.034
Knop JK, Hansen RG (1970) Uridine diphosphate glucose pyrophosphorylase. IV. Crystallization and properties of the enzyme from human liver. J Biol Chem 245(10):2499–2504. https://doi.org/10.1016/S0021-9258(18)63098-4
Kyryliv BY, Hunchak AV, Stefanyshyn OM (2018) Activity of hydrolytic enzyme in the poultry of different species. Scientific Messenger of LNU of Veterinary Medicine and Biotechnologies. Series: Agricultural Sciences 20(89):95–99. https://doi.org/10.32718/nvlvet8918
Lee YJ, Ji YR, Lee S, Choi MJ, Cho Y (2019) Microencapsulation of probiotic Lactobacillus acidophilus KBL409 by extrusion technology to enhance survival under simulated intestinal and freeze-drying conditions. J Microbiol Biotechnol. https://doi.org/10.4014/jmb.1903.03018
Li H, Li J, Jiao X, Li KH, Sun YJ, Zhou WL, Shen YL, Qian J, Chang AP, Wang JQ, Zhu H (2019) Characterization of the biosynthetic pathway of nucleotide sugar precursor UDP-glucose during sphingan WL gum production in Sphingomonas sp. WG-ScienceDirect Journal of Biotechnology 302:1–9. https://doi.org/10.1016/j.jbiotec.2019.06.005
Li B, Tian F, Liu X, Zhao J, Hao Z, Wei C (2011) Effects of cryoprotectants on viability of Lactobacillus reuteri CICC6226. Appl Microbiol Biotechnol 92(3):609–616. https://doi.org/10.1007/s00253-011-3269-4
Li HF, Zhang YY, Gao Y, Lan YY, Yin XP, Huang LF (2016) Characterization of UGPase from Aureobasidium pullulans NRRL Y-12974 and application in enhanced pullulan production. Applied Biochemistry and Biotechnology:1141–1153. https://doi.org/10.1007/s12010-015-1934-2
Luo SL, Dang LZ, Zhang KQ, Liang LM, Li GH (2015) Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett Appl Microbiol 60(1):72–78. https://doi.org/10.1111/lam.12339
Martz F, Wilczynska M, Kleczkowski LA (2002) Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochemical Journal 367(1):295–300. https://doi.org/10.1042/bj20020772
Munch-Petersen A, Kalckar HM, Cutolo E, Smith EEB (1953) Uridyl transferases and the formation of uridine triphosphate:enzymic production of uridine triphosphate: uridine diphosphoglucose pyrophosphorolysis. Nature 172(4388):1036–1037. https://doi.org/10.1038/1721036a0
Raimi OG, Hurtado-Guerrero R, Borodkin V, Ferenbach A, Urbaniak MD, Ferguson MAJ, van Aalten DMF (2020) A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor. RSC chemical biology 1:13–25. https://doi.org/10.1039/C9CB00017H
Rekik H, Jaouadi NZ, Gargouri F, Bejar W, Jaouadi B (2018) Production, purification and biochemical characterization of a novel detergent-stable serine alkaline protease from Bacillus safensis strain RH12. Int J Biol Macromol 121:1227–1239. https://doi.org/10.1016/j.ijbiomac.2018.10.139
Sharma M, Sharma S, Ray P, Chakraborti A (2020) Targeting Streptococcus pneumoniae UDP-glucose pyrophosphorylase (UGPase): in vitro validation of a putative inhibitor. Drug Target Insights 14(1):26–33. https://doi.org/10.33393/dti.2020.2103
Shi K, An W, Meng Q, Gu YJ, Liu SW (2021) Partial characterization and lyoprotective activity of exopolysaccharide from Oenococcus oeni 28A–1. Process Biochem 101(2):128–136. https://doi.org/10.1016/j.procbio.2020.10.015
Sun X, Zhang CY, Wu MY, Fan ZH, Liu SN, Zhu WB, Xiao DG (2016) MAL62 overexpression and NTH1 deletion enhance the freezing tolerance and fermentation capacity of the baker’s yeast in lean dough. Microbial Cell Factories 15(1). https://doi.org/10.1186/s12934-016-0453-3
Tamboli FA, More HN, Bhandugare SS, Patil AS, Jadhav NR, Killedar SG (2020) Estimation of total carbohydrate content by phenol sulphuric acid method from Eichhornia crassipes (Mart.) Solms. Asian Journal of Research in Chemistry 13(5):357–359. https://doi.org/10.5958/0974-4150.2020.00067.X
Tan H, Dong J, Wang G, Xu H, Zhang C, Xiao D (2014) Enhanced freeze tolerance of baker’s yeast by overexpressed trehalose-6-phosphate synthase gene (TPS1) and deleted trehalase genes in frozen dough. J Ind Microbiol Biotechnol 41(8):1275–1285. https://doi.org/10.1007/s10295-014-1467-7
Thoden JB, Holden HM (2007) Active site geometry of glucose-1-phosphate uridylyltransferase. Protein Sci 16(7):1379–1388. https://doi.org/10.1110/ps.072864707
Toit ED, Vesterlund S, Gueimonde M, Salminen S (2013) Assessment of the effect of stress-tolerance acquisition on some basic characteristics of specific probiotics. Int J Food Microbiol 165(1):51–61. https://doi.org/10.1016/j.ijfoodmicro.2013.04.022
Turnquist RL, Gillett TA, Hansen RG (1974) Uridine diphosphate glucose pyrophosphorylase. Crystallization and properties of the enzyme from rabbit liver and species comparisons. J Biol Chem 249(23):7695–7700. https://doi.org/10.1016/S0021-9258(19)81292-9
Velly H, Bouix M, Passot S, Penicaud C, Beinsteiner H, Ghorbal S, Lieben P, Fonseca F (2015) Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp. lactis TOMSC161. Appl Microbiol Biotechnol 99(2):907–918. https://doi.org/10.1007/s00253-014-6152-2
Villafranca JJ, Nowak T (1992) 2 metal ions at enzyme active sites. Enzymes 20:63–94. https://doi.org/10.1016/S1874-6047(08)60020-7
Wan RL, Sun J, He T, Hu YD, Zhao Y, Wu Y, Chun Z (2017) Cloning cDNA and functional characterization of UDP-glucose pyrophosphorylase in Dendrobium officinale. Biol Plant 61(1):147–154. https://doi.org/10.1007/s10535-016-0645-z
Wang Q, Zhang X, Li F, Hou Y, Liu X, Zhang X (2011) Identification of a UDP-glucose pyrophosphorylase from cotton (Gossypium hirsutum L.) involved in cellulose biosynthesis in Arabidopsis thaliana. Plant Cell Reports 30(7):1303–1312. https://doi.org/10.1007/s00299-011-1042-x
Wang X, Shao C, Liu L, Guo X, Xu Y, Lu X (2017) Optimization, partial characterization and antioxidant activity of an exopolysaccharide from Lactobacillus plantarum KX041. Int J Biol Macromol 103:1173–1184. https://doi.org/10.1016/j.ijbiomac.2017.05.118
Winter H, Huber SC (2010) Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci 19(1):31–67. https://doi.org/10.1080/07352680091139178
Wu C, Zhang J, Du G, Chen J (2013) Heterologous expression of Lactobacillus casei RecO improved the multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 during salt stress. Biores Technol 143:238–241. https://doi.org/10.1016/j.biortech.2013.05.050
Wu CH, Rismondo J, Morgan RML, Shen Y, Loessner MJ, Larrouy-Maumus G, Freemont PS, Gründling A (2021) Bacillus subtilis YngB contributes to wall teichoic acid glucosylation and glycolipid formation during anaerobic growth. J Biol Chem. https://doi.org/10.1016/j.jbc.2021.100384
Zannini E, Waters DM, Coffey A, Arendt EK (2016) Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl Microbiol Biotechnol 100(3):1121–1135. https://doi.org/10.1007/s00253-015-7172-2
Zayed G, Roos YH (2004) Influence of trehalose and moisture concentration on survival of Lactobacillus salivarius subjected to freeze-drying and storage. Process Biochem 39(9):1081–1086. https://doi.org/10.1016/s0032-9592(03)00222-x
Zhang K, Wu ZD, Tang DB, Luo K, Lu HX, Liu YY, Dong J, Wang X, Lv CW, Wang JC, Lu K (2017) Comparative transcriptome analysis reveals critical function of sucrose metabolism related-enzymes in starch accumulation in the storage root of sweet potato. Frontiers in Plant Science 8. https://doi.org/10.3389/fpls.2017.00914
Zhen N, Zeng XQ, Wang HJ, Yu J, Guo YX (2020) Effects of heat shock treatment on the survival rate of Lactobacillus acidophilus after freeze-drying. Food Res Int 136:1–10. https://doi.org/10.1016/j.foodres.2020.109507
Zhou YC, Bu ZY, Qian J, Chen YN, Qiao LJ, Yang S, Chen SP, Wang XL, Ren LZ, Yang YL (2020) The UTP-glucose-1-phosphate uridylyltransferase of Brucella melitensis inhibits the activation of NF-κB via regulating the bacterial type IV secretion system. Int J Biol Macromol 164:3098–3104. https://doi.org/10.1016/j.ijbiomac.2020.08.134
Zhuang Z, Yang G, Mai Q, Guo J, Zhuang L (2020) Physiological potential of extracellular polysaccharide in promoting Geobacter biofilm formation and extracellular electron transfer. Sci Total Environ 741:140365. https://doi.org/10.1016/j.scitotenv.2020.140365
Funding
This study received financial support from the National Natural Science Foundation of China (32072195, 41406165, 41641052, 31972093), the Science and Technology Department of Zhejiang Province (2019C02085), and the Education Department of Zhejiang Province (Y202148155, Y202148164, Y202148135).
Author information
Authors and Affiliations
Contributions
NZ and XQZ designed the study and performed the experiments; CYY and QYS prepared the manuscript and analyzed the data; XQZ reviewed and designed the manuscript; DDP, ZW, YXG, and ZDC helped improve the research plan; and CYY and QYS contributed to the revision of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Consent for publication
All authors revised and consented to publication of the findings.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhen, N., Ye, C., Shen, Q. et al. Heterologous expression and biological characteristics of UGPases from Lactobacillus acidophilus. Appl Microbiol Biotechnol 106, 2481–2491 (2022). https://doi.org/10.1007/s00253-022-11856-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11856-8