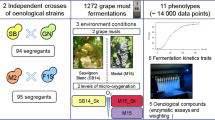
Abstract
The two most commonly used wine microorganisms, Saccharomyces cerevisiae yeast and Oenococcus oeni bacteria, are responsible for completion of alcoholic and malolactic fermentation (MLF), respectively. For successful co-inoculation, S. cerevisiae and O. oeni must be able to complete fermentation; however, this relies on compatibility between yeast and bacterial strains. For the first time, quantitative trait loci (QTL) analysis was used to elucidate whether S. cerevisiae genetic makeup can play a role in the ability of O. oeni to complete MLF. Assessment of 67 progeny from a hybrid S. cerevisiae strain (SBxGN), co-inoculated with a single O. oeni strain, SB3, revealed a major QTL linked to MLF completion by O. oeni. This QTL encompassed a well-known translocation, XV-t-XVI, that results in increased SSU1 expression and is functionally linked with numerous phenotypes including lag phase duration and sulphite export and production. A reciprocal hemizygosity assay was performed to elucidate the effect of the gene SSU1 in the SBxGN background. Our results revealed a strong effect of SSU1 haploinsufficiency on O. oeni’s ability to complete malolactic fermentation during co-inoculation and pave the way for the implementation of QTL mapping projects for deciphering the genetic bases of microbial interactions.
Key points
• For the first time, QTL analysis has been used to study yeast-bacteria interactions.
• A QTL encompassing a translocation, XV-t-XVI, was linked to MLF outcomes.
• S. cerevisiae SSU1 haploinsufficiency positively impacted MLF by O. oeni.




Similar content being viewed by others
Data availability
Raw data may be supplied upon request, at the discretion of the corresponding authors.
References
Abrahamse CE, Bartowsky EJ (2012) Timing of malolactic fermentation inoculation in Shiraz grape must and wine: influence on chemical composition. World J Microbiol Biotechnol 28:255–265. https://doi.org/10.1007/s11274-011-0814-3
Alexandre H, Costello PJ, Remize F, Guzzo J, Guilloux-Benatier M (2004) Saccharomyces cerevisiae–Oenococcus oeni interactions in wine: current knowledge and perspectives. Int J Food Microbiol 93:141–154. https://doi.org/10.1016/j.ijfoodmicro.2003.10.013
Antalick G, Perello MC, de Revel G (2013) Co-inoculation with yeast and LAB under winery conditions: modification of the aromatic profile of merlot wines. S Afr J Enol Vitic 34:223–232. https://doi.org/10.21548/34-2-1098
Arnink K, Henick-Kling T (2005) Influence of Saccharomyces cerevisiae and Oenococcus oeni strains on successful malolactic conversion in wine. Am J Enol Vitic 56:228–237
Atanassova M, Choiset Y, Dalgalarrondo M, Chobert JM, Dousset X, Ivanova I, Haertlé T (2003) Isolation and partial biochemical characterization of a proteinaceous anti-bacteria and anti-yeast compound produced by Lactobacillus paracasei subsp. paracasei strain M3. Int J Food Microbiol 87:63–73. https://doi.org/10.1016/S0168-1605(03)00054-0
Bartle L, Sumby K, Sundstrom J, Jiranek V (2019) The microbial challenge of winemaking: yeast-bacteria compatibility. FEMS Yeast Res 19:foz040. https://doi.org/10.1093/femsyr/foz040
Bartle L, Mitchell JG, Paterson JS (2020) Evaluating the cytometric detection and enumeration of the wine bacterium, Oenococcus oeni. Cytom Part A 99:399–406. https://doi.org/10.1002/cyto.a.24258
Bokulich NA, Bamforth CW (2013) The microbiology of malting and brewing. Microbiol Mol Biol Rev 77:157–172. https://doi.org/10.1128/MMBR.00060-12
Brice C, Sanchez I, Bigey F, Legras JL, Blondin B (2014) A genetic approach of wine yeast fermentation capacity in nitrogen-starvation reveals the key role of nitrogen signaling. BMC Genomics 15:495. https://doi.org/10.1186/1471-2164-15-495
Broman KW, Wu H, Sen Ś, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890. https://doi.org/10.1093/bioinformatics/btg112
Cañas PMI, Pérez-Martín F, Romero EG, Prieto SS, Herreros MDLLP (2012) Influence of inoculation time of an autochthonous selected malolactic bacterium on volatile and sensory profile of Tempranillo and Merlot wines. Int J Food Microbiol 156:245–254. https://doi.org/10.1016/j.ijfoodmicro.2012.03.033
Cañas PMI, Romero EG, Pérez-Martín F, Seseña S, Palop ML (2015) Sequential inoculation versus co-inoculation in Cabernet Franc wine fermentation. Food Sci Technol Int 21:203–212. https://doi.org/10.1177/1082013214524585
Capucho I, San Romão MV (1994) Effect of ethanol and fatty acids on malolactic activity of Leuconostoc oenos. Appl Microbiol Biotechnol 42:391–395. https://doi.org/10.1007/BF00902747
Carrete R, Vidal MT, Bordons A, Constantí M (2002) Inhibitory effect of sulfur dioxide and other stress compounds in wine on the ATPase activity of Oenococcus oeni. FEMS Microbiol Lett 211:155–159. https://doi.org/10.1111/j.1574-6968.2002.tb11218.x
Chasseriaud L, Coulon J, Marullo P, Albertin W, Bely M (2018) New oenological practice to promote non-Saccharomyces species of interest: saturating grape juice with carbon dioxide. Appl Microbiol Biotechnol 102:3779–3791. https://doi.org/10.1007/s00253-018-8861-4
Chen X, Niks RE, Hedley PE, Morris J, Druka A, Marcel TC, Vels A, Waugh R (2010) Differential gene expression in nearly isogenic lines with QTL for partial resistance to Puccinia hordei in barley. BMC Genomics 11:629. https://doi.org/10.1186/1471-2164-11-629
Cheraiti N, Guezenec S, Salmon JM (2010) Very early acetaldehyde production by industrial Saccharomyces cerevisiae strains: a new intrinsic character. Appl Microbiol Biotechnol 86:693-700. https://doi.org/10.1007/s00253-009-2337-5
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Comitini F, Ciani M (2007) The inhibitory activity of wine yeast starters on malolactic bacteria. Ann Microbiol 57:61–66. https://doi.org/10.1007/BF03175051
Comitini F, Ferretti R, Clementi F, Mannazzu I, Ciani M (2005) Interactions between Saccharomyces cerevisiae and malolactic bacteria: preliminary characterization of a yeast proteinaceous compound (s) active against Oenococcus oeni. J Appl Microbiol 99:105–111. https://doi.org/10.1111/j.1365-2672.2005.02579.x
Decroocq V, Foulongne M, Lambert P, Gall OL, Mantin C, Pascal T, Schurdi-Levraud V, Kervella J (2005) Analogues of virus resistance genes map to QTLs for resistance to sharka disease in Prunus davidiana. Mol Gen Genomics 272:680–689. https://doi.org/10.1007/s00438-004-1099-0
Delcourt F, Taillandier P, Vidal F, Strehaiano P (1995) Influence of pH, malic acid and glucose concentrations on malic acid consumption by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 43:321–324. https://doi.org/10.1007/BF00172832
Divol B, Du Toit M, Duckitt E (2012) Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl Microbiol Biotechnol 95:601–613. https://doi.org/10.1007/s00253-012-4186-x
Eder M, Sanchez I, Brice C, Camarasa C, Legras JL, Dequin S (2018) QTL mapping of volatile compound production in Saccharomyces cerevisiae during alcoholic fermentation. BMC Genomics 19:166. https://doi.org/10.1186/s12864-018-4562-8
El-Gendy SM, Abdel-Galil H, Shahin Y, Hegazi FZ (1983) Acetoin and diacetyl production by homo- and heterofermentative lactic acid bacteria. J Food Prot 46:420–425. https://doi.org/10.4315/0362-028X-46.5.420
Eun MH, Han JH, Yoon JB, Lee J (2016) QTL mapping of resistance to the Cucumber mosaic virus P1 strain in pepper using a genotyping-by-sequencing analysis. Hortic Environ Biotechnol 57:589–597. https://doi.org/10.1007/s13580-016-0128-3
Gao C, Fleet GH (1995) Degradation of malic and tartaric acids by high density cell suspensions of wine yeasts. Food Microbiol 12:65–71. https://doi.org/10.1016/S0740-0020(95)80080-8
García-Ríos E, Guillamón JM (2019) Sulfur dioxide resistance in Saccharomyces cerevisiae: beyond SSU1. Microb Cell 6:527–530. https://doi.org/10.15698/mic2019.12.699
García-Ríos E, Nuévalos M, Barrio E, Puig S, Guillamón JM (2019) A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ Microbiol 21:1771–1781. https://doi.org/10.1111/1462-2920.14586
Guzzo J, Jobin MP, Delmas F, Fortier LC, Garmyn D, Tourdot-Maréchal R, Lee B, Diviès C (2000) Regulation of stress response in Oenococcus oeni as a function of environmental changes and growth phase. Int J Food Microbiol 55:27–31. https://doi.org/10.1016/S0168-1605(00)00209-9
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324. https://doi.org/10.1038/hdy.1992.131
Hranilovic A, Gambetta JM, Schmidtke L, Boss PK, Grbin PR, Masneuf-Pomarede I, Bely M, Albertin W, Jiranek V (2018) Oenological traits of Lachancea thermotolerans show signs of domestication and allopatric differentiation. Sci Rep 8:1–13. https://doi.org/10.1038/s41598-018-33105-7
Huang C, Roncoroni M, Gardner RC (2014) MET2 affects production of hydrogen sulfide during wine fermentation. Appl Microbiol Biotechnol 98:7125–7135. https://doi.org/10.1007/s00253-014-5789-1
Jara M, Cubillos FA, García V, Salinas F, Aguilera O, Liti G, Martínez C (2014) Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS One 9:e86533. https://doi.org/10.1371/journal.pone.0086533
Jarosz DF, Brown JC, Walker GA, Datta MS, Ung WL, Lancaster AK, Rotem A, Chang A, Newby GA, Weitz DA, Bisson LF (2014) Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158:1083–1093. https://doi.org/10.1016/j.cell.2014.07.025
Knoll C, Fritsch S, Schnell S, Grossmann M, Krieger-Weber S, Du Toit M, Rauhut D (2012) Impact of different malolactic fermentation inoculation scenarios on Riesling wine aroma. World J Microbiol Biotechnol 28:1143–1153. https://doi.org/10.1007/s11274-011-0917-x
Liti G, Louis EJ (2012) Advances in quantitative trait analysis in yeast. PLoS Genet 8:e1002912. https://doi.org/10.1371/journal.pgen.1002912
Liu SQ, Pilone GJ (2000) An overview of formation and roles of acetaldehyde in winemaking with emphasis on microbiological implications. Int J Food Sci Technol 35:49–61. https://doi.org/10.1046/j.1365-2621.2000.00341.x
Martí-Raga M, Peltier E, Mas A, Beltran G, Marullo P (2017) Genetic causes of phenotypic adaptation to the second fermentation of sparkling wines in Saccharomyces cerevisiae. G3 Genes Genom Genet 7:399–412. https://doi.org/10.1534/g3.116.037283
Marullo P, Yvert G, Bely M, Aigle M, Dubourdieu D (2007a) Efficient use of DNA molecular markers to construct industrial yeast strains. FEMS Yeast Res 7:1295–1306. https://doi.org/10.1111/j.1567-1364.2007.00281.x
Marullo P, Aigle M, Bely M, Masneuf-Pomarede I, Durrens P, Dubourdieu D, Yvert G (2007b) Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res 7:941–952. https://doi.org/10.1111/j.1567-1364.2007.00252.x
Marullo P, Durrens P, Peltier E, Bernard M, Mansour C, Dubourdieu D (2019) Natural allelic variations of Saccharomyces cerevisiae impact stuck fermentation due to the combined effect of ethanol and temperature; a QTL-mapping study. BMC Genomics 20:680. https://doi.org/10.1186/s12864-019-5959-8
Marullo P, Claisse O, Eder ML, Börlin M, Feghali N, Bernard M, Legras JL, Albertin W, Rosa AL, Masneuf-Pomarede I (2020) SSU1 checkup, a rapid tool for detecting chromosomal rearrangements related to the SSU1 promoter in Saccharomyces cerevisiae: an ecological and technological study on wine yeast. Front Microbiol 11:1331. https://doi.org/10.3389/fmicb.2020.01331
Mendoza LM, de Nadra MCM, Farías ME (2010) Antagonistic interaction between yeasts and lactic acid bacteria of oenological relevance: partial characterization of inhibitory compounds produced by yeasts. Food Res Int 43:1990–1998. https://doi.org/10.1016/j.foodres.2010.05.017
Mounier J, Monnet C, Vallaeys T, Arditi R, Sarthou AS, Hélias A, Irlinger F (2008) Microbial interactions within a cheese microbial community. Appl Environ Microbiol 74:172–181. https://doi.org/10.1128/AEM.01338-07
Muñoz V, Beccaria B, Abreo E (2014) Simultaneous and successive inoculations of yeasts and lactic acid bacteria on the fermentation of an unsulfited Tannat grape must. Braz J Microbiol 45:59–66. https://doi.org/10.1590/S1517-83822014000100009
Nehme N, Mathieu F, Taillandier P (2008) Quantitative study of interactions between Saccharomyces cerevisiae and Oenococcus oeni strains. J Ind Microbiol Biotechnol 35:685–693. https://doi.org/10.1007/s10295-008-0328-7
Nehme N, Mathieu F, Taillandier P (2010) Impact of the co-culture of Saccharomyces cerevisiae–Oenococcus oeni on malolactic fermentation and partial characterization of a yeast-derived inhibitory peptidic fraction. Food Microbiol 27:150–157. https://doi.org/10.1016/j.fm.2009.09.008
Nissen P, Nielsen D, Arneborg N (2003) Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell–cell contact-mediated mechanism. Yeast 20:331–341. https://doi.org/10.1002/yea.965
Nissen P, Nielsen D, Arneborg N (2004) The relative glucose uptake abilities of non-Saccharomyces yeasts play a role in their coexistence with Saccharomyces cerevisiae in mixed cultures. Appl Microbiol Biotechnol 64:543–550. https://doi.org/10.1007/s00253-003-1487-0
Osborne JP, Edwards CG (2006) Inhibition of malolactic fermentation by Saccharomyces during alcoholic fermentation under low- and high-nitrogen conditions: a study in synthetic media. Aust J Grape Wine Res 12:69–78. https://doi.org/10.1111/j.1755-0238.2006.tb00045.x
Park H, Bakalinsky AT (2000) SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16:881–888. https://doi.org/10.1002/1097-0061(200007)16:10<881::AID-YEA576>3.0.CO;2-3
Peltier E, Bernard M, Trujillo M, Prodhomme D, Barbe JC, Gibon Y, Marullo P (2018a) Wine yeast phenomics: a standardized fermentation method for assessing quantitative traits of Saccharomyces cerevisiae strains in enological conditions. PLoS One 13:e0190094. https://doi.org/10.1371/journal.pone.0190094
Peltier E, Sharma V, Raga MM, Roncoroni M, Bernard M, Jiranek V, Gibon Y, Marullo P (2018b) Dissection of the molecular bases of genotype x environment interactions: a study of phenotypic plasticity of Saccharomyces cerevisiae in grape juices. BMC Genomics 19:772. https://doi.org/10.1186/s12864-018-5145-4
Peltier E, Friedrich A, Schacherer J, Marullo P (2019) Quantitative trait nucleotides impacting the technological performances of industrial Saccharomyces cerevisiae strains. Front Genet 10:683. https://doi.org/10.3389/fgene.2019.00683
Pérez-Ortín JE, Querol A, Puig S, Barrio E (2002) Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12:1533–1539. https://doi.org/10.1101/gr.436602
R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/. Accessed 4 Sept 2017
Renault P, Miot-Sertier C, Marullo P, Hernández-Orte P, Lagarrigue L, Lonvaud-Funel A, Bely M (2009) Genetic characterization and phenotypic variability in Torulaspora delbrueckii species: potential applications in the wine industry. Int J Food Microbiol 134:201–210. https://doi.org/10.1016/j.ijfoodmicro.2009.06.008
Renault PE, Albertin W, Bely M (2013) An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl Microbiol Biotechnol 97:4105–4119. https://doi.org/10.1007/s00253-012-4660-5
Renault P, Coulon J, de Revel G, Barbe JC, Bely M (2015) Increase of fruity aroma during mixed T. delbrueckii/S. cerevisiae wine fermentation is linked to specific esters enhancement. Int J Food Microbiol 207:40–48. https://doi.org/10.1016/j.ijfoodmicro.2015.04.037
Renault P, Coulon J, Moine V, Thibon C, Bely M (2016) Enhanced 3-sulfanylhexan-1-ol production in sequential mixed fermentation with Torulaspora delbrueckii/Saccharomyces cerevisiae reveals a situation of synergistic interaction between two industrial strains. Front Microbiol 7:293. https://doi.org/10.3389/fmicb.2016.00293
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75:149–164. https://doi.org/10.1007/s00253-006-0798-3
Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A (2006) Handbook of enology, Vol. 1: The microbiology of wine and vinifications, 2nd edn. Wiley, doi:https://doi.org/10.1002/0470010363
Roncoroni M, Santiago M, Hooks DO, Moroney S, Harsch MJ, Lee SA, Richards KD, Nicolau L, Gardner RC (2011) The yeast IRC7 gene encodes a β-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol 28:926–935. https://doi.org/10.1016/j.fm.2011.01.002
Rossouw D, Du Toit M, Bauer FF (2012) The impact of co-inoculation with Oenococcus oeni on the trancriptome of Saccharomyces cerevisiae and on the flavour-active metabolite profiles during fermentation in synthetic must. Food Microbiol 29:121–131. https://doi.org/10.1016/j.fm.2011.09.006
Sadoudi M, Tourdot-Maréchal R, Rousseaux S, Steyer D, Gallardo-Chacón JJ, Ballester J, Vichi S, Guérin-Schneider R, Caixach J, Alexandre H (2012) Yeast–yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol 32:243–253. https://doi.org/10.1016/j.fm.2012.06.006
Salinas F, Cubillos FA, Soto D, Garcia V, Bergström A, Warringer J, Ganga MA, Louis EJ, Liti G, Martinez C (2012) The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS One 7:e49640. https://doi.org/10.1371/journal.pone.0049640
Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW (2002a) Dissecting the architecture of a quantitative trait locus in yeast. Nature 416:326–330. https://doi.org/10.1038/416326a
Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW (2002b) Systematic screen for human disease genes in yeast. Nat Genet 31:400–404. https://doi.org/10.1038/ng929
Steyer D, Ambroset C, Brion C, Claudel P, Delobel P, Sanchez I, Erny C, Blondin B, Karst F, Legras JL (2012) QTL mapping of the production of wine aroma compounds by yeast. BMC Genomics 13:573. https://doi.org/10.1186/1471-2164-13-573
Tempère S, Marchal A, Barbe JC, Bely M, Masneuf-Pomarede I, Marullo P, Albertin W (2018) The complexity of wine: clarifying the role of microorganisms. Appl Microbiol Biotechnol 102:3995–4007. https://doi.org/10.1007/s00253-018-8914-8
Treu L, Toniolo C, Nadai C, Sardu A, Giacomini A, Corich V, Campanaro S (2014) The impact of genomic variability on gene expression in environmental Saccharomyces cerevisiae strains. Environ Microbiol 16:1378–1397. https://doi.org/10.1111/1462-2920.12327
Tristezza M, di Feo L, Tufariello M, Grieco F, Capozzi V, Spano G, Mita G (2016) Simultaneous inoculation of yeasts and lactic acid bacteria: effects on fermentation dynamics and chemical composition of Negroamaro wine. LWT-Food Sci Technol 66:406–412. https://doi.org/10.1016/j.lwt.2015.10.064
Walker ME, Nguyen TD, Liccioli T, Schmid F, Kalatzis N, Sundstrom JF, Gardner JM, Jiranek V (2014) Genome-wide identification of the Fermentome; genes required for successful and timely completion of wine-like fermentation by Saccharomyces cerevisiae. BMC Genomics 15:552. https://doi.org/10.1186/1471-2164-15-552
Yéramian N, Chaya C, Suárez Lepe JA (2007) L-(-)-malic acid production by Saccharomyces spp. during the alcoholic fermentation of wine. J Agric Food Chem 55:912–919. https://doi.org/10.1021/jf061990w
Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P (2014) QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One 9:e86298. https://doi.org/10.1371/journal.pone.0086298
Acknowledgements
We thank Hélène Mesnage for her assistance in performing the QTL fermentation experiment. We also acknowledge Nick van Holst Pellekaan for capturing images of the automated fermentation platform.
Funding
This work was supported by Australia’s grape growers and winemakers through their investment body, Wine Australia, with matching funds from the Australian Government. LB was supported by joint scholarships from The University of Adelaide and Wine Australia (AGW Ph 1510). JS was supported by Wine Australia project funding (UA1707). JS, KS and VJ are supported by The Australian Research Council Training Centre for Innovative Wine Production (www.ARCwinecentre.org.au; project number IC170100008), which is funded by the Australian Government with additional support from Wine Australia and industry partners. The University of Adelaide is a member of the Wine Innovation Cluster in Adelaide (http://www.thewaite.org/waite-partners/wine-innovation-cluster/). PM and EP are supported by Biolaffort (Laffort® Research & Development subsidiary) for this project; in addition PM received a grant from Aquitaine Region (Sesam Project) for genome sequencing and QTL analysis.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. LB carried out laboratory fermentation experiments; PM and EP provided yeast strains. Data analysis and interpretation was performed by LB, EP and PM. LB wrote the manuscript and EP, JS, KS, JGM, VJ and PM reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
EP and PM are employed by Biolaffort. This does not alter the authors’ adherence to all the journal policies on sharing data and materials. All other authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bartle, L., Peltier, E., Sundstrom, J.F. et al. QTL mapping: an innovative method for investigating the genetic determinism of yeast-bacteria interactions in wine. Appl Microbiol Biotechnol 105, 5053–5066 (2021). https://doi.org/10.1007/s00253-021-11376-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11376-x