Abstract
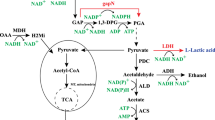
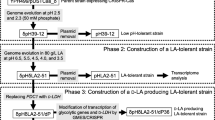
There is an increasing demand for microbial production of lactic acid (LA) as a monomer of biodegradable poly lactic acid (PLA). Both optical isomers, D-LA and L-LA, are required to produce stereocomplex PLA with improved properties. In this study, we developed Saccharomyces cerevisiae strains for efficient production of D-LA. D-LA production was achieved by expressing highly stereospecific D-lactate dehydrogenase gene (ldhA, LEUM_1756) from Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 in S. cerevisiae lacking natural LA production activity. D-LA consumption after glucose depletion was inhibited by deleting DLD1 encoding D-lactate dehydrogenase and JEN1 encoding monocarboxylate transporter. In addition, ethanol production was reduced by deleting PDC1 and ADH1 genes encoding major pyruvate decarboxylase and alcohol dehydrogenase, respectively, and glycerol production was eliminated by deleting GPD1 and GPD2 genes encoding glycerol-3-phosphate dehydrogenase. LA tolerance of the engineered D-LA-producing strain was enhanced by adaptive evolution and overexpression of HAA1 encoding a transcriptional activator involved in weak acid stress response, resulting in effective D-LA production up to 48.9 g/L without neutralization. In a flask fed-batch fermentation under neutralizing condition, our evolved strain produced 112.0 g/L D-LA with a yield of 0.80 g/g glucose and a productivity of 2.2 g/(L · h).






Similar content being viewed by others
References
Abbott DA, Suir E, van Maris AJ, Pronk JT (2008) Physiological and transcriptional responses to high concentrations of lactic acid in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 74(18):5759–5768
Abdel-Rahman MA, Tashiro Y, Sonomoto K (2013) Recent advances in lactic acid production by microbial fermentation processes. Biotechnol Adv 31(6):877–902
Branduardi P, Sauer M, De Gioia L, Zampella G, Valli M, Mattanovich D, Porro D (2006) Lactate production yield from engineered yeasts is dependent from the host background, the lactate dehydrogenase source and the lactate export. Microb Cell Fact 5(1):1–12
Cho BR, Lee P, Hahn JS (2014) CK2-dependent inhibitory phosphorylation is relieved by Ppt1 phosphatase for the ethanol stress-specific activation of Hsf1 in Saccharomyces cerevisiae. Mol Microbiol 93(2):306–316
Collart MA, Oliviero S (2001) Preparation of yeast RNA. Curr Protoc Mol Biol 13(13):12
Colombié S, Dequin S, Sablayrolles JM (2003) Control of lactate production by Saccharomyces cerevisiae expressing a bacterial LDH gene. Enzyme Microb Technol 33(1):38–46
Dato L, Berterame N, Ricci M, Paganoni P, Palmieri L, Porro D, Branduardi P (2014) Changes in SAM2 expression affect lactic acid tolerance and lactic acid production in Saccharomyces cerevisiae. Microb Cell Factories 13:147
Garlotta D (2001) A literature review of poly(lactic acid). J Polym Environ 9(2):63–84
Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30(6), e23
Hofvendahl K, Hahn-Hägerdal B (2000) Factors affecting the fermentative lactic acid production from renewable resources1. Enzyme Microb Technol 26(2–4):87–107
Ida Y, Hirasawa T, Furusawa C, Shimizu H (2013) Utilization of Saccharomyces cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for anaerobic bioproduction. Appl Microbiol Biotechnol 97(11):4811–4819
Ikada Y, Jamshidi K, Tsuji H, Hyon SH (1987) Stereocomplex formation between enantiomeric poly(lactides). Macromolecules 20(4):904–906
Inaba T, Watanabe D, Yoshiyama Y, Tanaka K, Ogawa J, Takagi H, Shimoi H, Shima J (2013) An organic acid-tolerant HAA1-overexpression mutant of an industrial bioethanol strain of Saccharomyces cerevisiae and its application to the production of bioethanol from sugarcane molasses. AMB express 3(1):74
Ishida N, Saitoh S, Tokuhiro K, Nagamori E, Matsuyama T, Kitamoto K, Takahashi H (2005) Efficient production of L-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated L-lactate dehydrogenase gene. Appl Environ Microbiol 71(4):1964–1970
Ishida N, Saitoh S, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2006a) The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci, Biotechnol, Biochem 70(5):1148–1153
Ishida N, Suzuki T, Tokuhiro K, Nagamori E, Onishi T, Saitoh S, Kitamoto K, Takahashi H (2006b) D-lactic acid production by metabolically engineered Saccharomyces cerevisiae. J Biosci Bioeng 101(2):172–177
Jun C, Sa YS, Gu S-A, Joo JC, Kim S, Kim K-J, Kim YH (2013) Discovery and characterization of a thermostable D-lactate dehydrogenase from Lactobacillus jensenii through genome mining. Process Biochem 48(1):109–117
Kawahata M, Masaki K, Fujii T, Iefuji H (2006) Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res 6(6):924–936
Kim S, Hahn J-S (2015) Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 31:94–101
Lee JY, Kang CD, Lee SH, Park YK, Cho KM (2015) Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid. Biotechnol Bioeng 112(4):751–758
Li L, Eom H-J, Park J-M, Seo E, Ahn JE, Kim T-J, Kim JH, Han NS (2012) Characterization of the major dehydrogenase related to D-lactic acid synthesis in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293. Enzyme Microb Technol 51(5):274–279
Lodi T, Alberti A, Guiard B, Ferrero I (1999) Regulation of the Saccharomyces cerevisiae DLD1 gene encoding the mitochondrial protein D-lactate ferricytochrome c oxidoreductase by HAP1 and HAP2/3/4/5. Mol Gen Genet 262(4–5):623–632
Lodi T, Fontanesi F, Guiard B (2002) Co-ordinate regulation of lactate metabolism genes in yeast: the role of the lactate permease gene JEN1. Mol Genet Genomics 266(5):838–847
Mira NP, Teixeira MC, Sá-Correia I (2010) Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS: J Integrative Biol 14(5):525–540
Mira NP, Henriques SF, Keller G, Teixeira MC, Matos RG, Arraiano CM, Winge DR, Sa-Correia I (2011) Identification of a DNA-binding site for the transcription factor Haa1, required for Saccharomyces cerevisiae response to acetic acid stress. Nucleic Acids Res 39(16):6896–907
Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156(1):119–122
Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008) Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78(3):449–454
Pacheco A, Talaia G, Sá-Pessoa J, Bessa D, Gonçalves MJ, Moreira R, Paiva S, Casal M, Queirós O (2012) Lactic acid production in Saccharomyces cerevisiae is modulated by expression of the monocarboxylate transporters Jen1 and Ady2. FEMS Yeast Res 12(3):375–381
Porro D, Brambilla L, Ranzi BM, Martegani E, Alberghina L (1995) Development of metabolically engineered Saccharomyces cerevisiae cells for the production of lactic acid. Biotechnol Prog 11(3):294–298
Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2005) Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl Environ Microbiol 71(5):2789–2792
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122(1):19–27
Skory C (2003) Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol 30(1):22–27
Sugiyama M, Akase S-P, Nakanishi R, Horie H, Kaneko Y, Harashima S (2014) Nuclear localization of Haa1, which is linked to its phosphorylation status, mediates lactic acid tolerance in Saccharomyces cerevisiae. Appl Environ Microbiol 80(11):3488–3495
Suzuki T, Sakamoto T, Sugiyama M, Ishida N, Kambe H, Obata S, Kaneko Y, Takahashi H, Harashima S (2013) Disruption of multiple genes whose deletion causes lactic-acid resistance improves lactic-acid resistance and productivity in Saccharomyces cerevisiae. J Biosci Bioeng 115(5):467–474
Tokuhiro K, Ishida N, Nagamori E, Saitoh S, Onishi T, Kondo A, Takahashi H (2009) Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl Microbiol Biotechnol 82(5):883–890
Vaidya AN, Pandey RA, Mudliar S, Kumar MS, Chakrabarti T, Devotta S (2005) Production and recovery of lactic acid for polylactide—an overview. Crit Rev Environ Sci Technol 35(5):429–467
Valli M, Sauer M, Branduardi P, Borth N, Porro D, Mattanovich D (2006) Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl Environ Microbiol 72(8):5492–5499
van Maris AJA, Geertman J-MA, Vermeulen A, Groothuizen MK, Winkler AA, Piper MDW, van Dijken JP, Pronk JT (2004a) Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microbiol 70(1):159–166
van Maris AJA, Winkler AA, Porro D, van Dijken JP, Pronk JT (2004b) Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microbiol 70(5):2898–2905
Acknowledgments
We are grateful to Dr. N. S. Han for providing a plasmid.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (NRF-2015R1A2A2A01005429).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Baek, SH., Kwon, E.Y., Kim, Y.H. et al. Metabolic engineering and adaptive evolution for efficient production of D-lactic acid in Saccharomyces cerevisiae . Appl Microbiol Biotechnol 100, 2737–2748 (2016). https://doi.org/10.1007/s00253-015-7174-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7174-0