Abstract
Porcine parvovirus type 1 (PPV1) is a major causative agent of embryonic and fetal death in swine. The PPV1 VP2 protein is closely associated with viral immunogenicity for eliciting neutralizing antibodies, but its antigenic structures have been largely unknown. We generated three monoclonal antibodies (MAbs) against baculovirus-expressed recombinant PPV1 VP2 protein. A PEPSCAN analysis identified the minimal B cell linear epitopes of PPV1 VP2 based on these MAbs. Three core epitopes, 228QQITDA233, 284RSLGLPPK291, and 344FEYSNGGPFLTPI356, were defined and mapped onto three-dimensional models of the PPV1 virion and VP2 monomer. The epitope 228QQITDA233 is exposed on the virion surface, and the other two are located inside the protein. An alignment of the PPV1 VP2 amino acid sequences showed that 284RSLGLPPK291 and 344FEYSNGGPFLTPI356 are absolutely conserved, whereas 228QQITDA233 has a single substitution at residue 233 in some (S → A or T). We developed a VP2 epitope-based indirect enzyme-linked immunosorbent assay (iELISA) to test for anti-PPV1 antibodies. In a comparative analysis with an immunoperoxidase monolayer assay using 135 guinea pig sera, the VP2-epitope-based iELISA had a concordance rate of 85.19 %, sensitivity of 83.33 %, and specificity of 85.47 %. MAb 8H6 was used to monitor VP2 during the PPV1 replication cycle in vitro with an indirect immunofluorescence assay, which indicated that newly encapsulated virions are released from the nucleus at 24 h postinfection and the PPV1 replication cycle takes less than 24 h. This study provides valuable information clarifying the antigenic structure of PPV1 VP2 and lays the foundations for PPV1 serodiagnosis and antigen detection.
Similar content being viewed by others
Introduction
Parvoviruses are small, single-stranded DNA viruses that are responsible for a number of diseases in various hosts. With the development of molecular detection technologies, several novel porcine parvoviruses (PPVs) have been identified in swine herds, including porcine parvovirus type 2 (PPV2), PPV3, PPV4, PPV5, and PPV6, and their prevalence and potential roles in clinical diseases have been investigated to some extent (Cheung et al. 2010; Hijikata et al. 2001; Ni et al. 2014; Sun et al. 2015; Wang et al. 2010). However, because the isolation of PPV2–6 is limited, their pathogenicity has not yet been clarified. The classical porcine parvovirus type 1 (PPV1) was first isolated in 1965 and is an etiological agent for reproductive disturbances in swine, leading to stillbirth, mummification, embryonic death, and infertility and has thus caused great economic losses for the swine industry around the world (Adlhoch et al. 2010; Mahnel et al. 1966; Opriessnig et al. 2004; Opriessnig et al. 2014).
PPV1, reclassified as a member of the genus Protoparvovirus, belongs to the subfamily Parvovirinae, in the family Parvoviridae (Cotmore et al. 2014). It has a genome of about 5 kb, which includes two main open reading frames (ORFs), ORF1 and ORF2. ORF1 is predominantly transcribed and translated into three nonstructural proteins (NS1, NS2, and NS3), which are responsible for viral infection (Nuesch et al. 1998; Nuesch and Rommelaere 2006; Rhode 1989). ORF2 primarily encodes three structural proteins (VP1, VP2, and VP3), which determine viral fitness in different host cell lines (Fernandes et al. 2011). The amino acid sequence of the VP2 protein is 150 amino acids shorter than that of the VP1 protein at the N-terminus as the result of alternative RNA splicing, and the VP3 protein is formed by the proteolytic cleavage of the VP2 protein at the N-terminus (Bergeron et al. 1993). Of the three structural proteins, VP2 is the major capsid protein. It automatically assembles into virus-like particles (VLPs) and contains a novel nuclear localization motif, which is critical for the assembly of newly virions during viral proliferation (Boisvert et al. 2014; Maranga et al. 2003). Most importantly, the PPV1 VP2 protein plays a dominant role in eliciting neutralizing antibodies against viral infection (Antonis et al. 2006; Rueda et al. 2000; Xu and Li 2007). Because its immunoprotective importance, we require more information about its antigenic changes in response to selective pressures. However, its antigenic structures have been largely unknown, even though many capsid sequences of various antigenic variants are available. Until now, as for PPV1 antigenic structures, only one mimotope of the VP1 protein has been identified with a phage display technique, and the general antigenic composition has been roughly scanned with polyclonal antisera and synthetic peptides (Kamstrup et al. 1998; Xie et al. 2010). Because there have been no updates on the antigenic structures of PPV1 VP2 for the last 18 years, it is crucial to determine how these epitopes have changed. The analysis of antigenic sites might also facilitate the development of effective diagnostic tools for clinical practice (Hu et al. 2014).
Here, we expressed the PPV1 VP2 protein in a baculovirus expression system, generated three PPV1 VP2 protein-specific monoclonal antibodies (MAbs) by an immunoperoxidase monolayer assay (IPMA), and identified their corresponding core epitopes. A VP2 epitope-based indirect enzyme-linked immunosorbent assay (iELISA) was established and evaluated for the serodiagnosis of anti-PPV1 antibodies in guinea pig sera, and a MAb-based indirect immunofluorescence assay (IFA) was established for monitoring of PPV1 VP2 protein during viral replication. This work provides valuable insight into the antigenic structures of PPV1 VP2, extending our understanding of the mechanism of antigenic drift, and will contribute to the establishment of diagnostic methods for PPV1.
Materials and methods
Virus, plasmids, and cells
The PPV1-LH strain was recently isolated from a pig with reproductive failure, and the nucleotide sequence of its VP2 protein has been submitted to the GenBank database under accession number KP245935. The baculovirus transfer vector pFastBac™ Dual (Invitrogen, Carlsbad, CA, USA) and the prokaryotic expression vector pGEX-6P-1 (GE Healthcare, Uppsala, Sweden) were used in this study to express the complete and truncated PPV1 VP2 proteins, respectively. The myeloma cell line SP2/0 and swine testicle (ST) cell line were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen) in a humidified incubator with 5 % CO2 at 37 °C. Spodoptera frugiperda (Sf21) insect cell line was cultured in Grace’s Insect Medium (Invitrogen) at 27 °C. All culture media were supplemented with 10 % fetal bovine serum (Invitrogen) and antibiotics (0.1 mg/mL streptomycin and 100 IU/mL penicillin).
Sera
Twenty-four specific pathogen-free Hartley guinea pigs (5 weeks old) were bought from Vital River Laboratories (Beijing, China) and used for the immunization test. Guinea pigs 1–3 were injected intramuscularly with an inactivated PPV1 vaccine at 0.5 mL/dose (0.5 × 107 TCID50); guinea pigs 4–15 were injected with 300 μg of baculovirus-expressed PPV1 VP2 protein emulsified with the oil-in-water adjuvant ISA 15A VG (Seppic, Castres, France) in a ratio of 85:15 (v/v); guinea pigs 16–24 received an injection of the mock inoculum and acted as the mock control group. These immunization regimens were repeated 3 weeks later. Blood samples were taken weekly, and the sera were isolated and tested with the IPMA.
Construction of recombinant plasmid
The amplification and purification of the PPV1 VP2-encoding gene have been described previously (Sun et al. 2015). After digestion with XbaI and PstI, the target gene was inserted into the multiple cloning site I of the pFastBac™ Dual transfer vector under the control of the polyhedrin (PPH) promoter. After the construct was confirmed with dual-enzyme digestion (XbaI + PstI) and sequencing, the recombinant transfer vector was designated pFBD-PPV1-VP2.
Recombinant protein expression and quantification
The recombinant baculovirus (referred to as rBACV-pFBD-PPV1-VP2) was produced with the Bac-to-Bac® baculovirus expression system (Invitrogen). Sf21 cells were infected with rBACV-pFBD-PPV1-VP2 and harvested at 72 h postinfection (hpi). The expressed protein samples were separated with 12 % SDS-PAGE and then transferred onto nitrocellulose (NC) membranes. A Western blotting analysis was performed with anti-PPV1 swine serum, and the target protein band was visualized with the 3,3′-diaminobenzidine (DAB) substrate (Zsgb-bio, Beijing, China). The PPV1 VP2 protein was quantified as previously described (Wang et al. 2013). In brief, the percentage (P%) of VP2 protein in the total protein was determined with a thin-layer chromatography scanner (Camag, Muttenz, Switzerland). The concentration of total protein (C T) was measured with a BCA protein assay kit (Pierce, Rockford, USA) using bovine serum albumin (BSA) as the standard. The concentration of VP2 protein was then calculated with the formula: CT × P% μg/mL.
Immunization of BALB/c mice and preparation of MAbs against PPV1 VP2 protein
Six- to eight-week-old female BALB/c mice were immunized subcutaneously with 100 μg of baculovirus-expressed recombinant PPV1 VP2 protein emulsified with Freund’s complete adjuvant and then with VP2 emulsified with Freund’s incomplete adjuvant (Sigma-Aldrich, St. Louis, USA) at an interval of 3 weeks. The mice were euthanized for hybridoma generation 3 days after a final booster immunization containing the same amount of recombinant VP2 protein. The mouse splenocytes were separated and fused with SP2/0 myeloma cells using polyethylene glycol (PEG) 1450 (Sigma-Aldrich). The hybridoma cells were then plated in 96-well plates in hypoxanthine–aminopterin–thymidine (HAT) selection medium, which was replaced with hypoxanthine–thymidine (HT) selection medium after 5 days (Wang et al. 2015). After HAT/HT selection, the hybridoma supernatants were screened with IPMA to test the PPV1-specific MAbs, as previously described (Liu et al. 2004). In brief, ST cells (80–90 % confluence) were inoculated with the PPV1-LH strain at a concentration of 104 TCID50/well and were fixed at 32 hpi with 33.3 % acetone at room temperature for 20 min and then dried. The hybridoma supernatants were added to the IPMA plates and incubated at 37 °C for 1 h. After the plates were washed three times with phosphate-buffered saline (PBS), a horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody (1:4000; Sigma-Aldrich) was added and incubated at 37 °C for 30 min. After the samples were washed, the color reaction was developed with 3-amino-9-ethylcarbazole and hydrogen peroxide in 0.05 M acetate buffer (pH 5.0) at 37 °C for 15 min. After the removal of the substrate, the IPMA plates were examined under an inverted light microscope. The positive hybridoma clones then underwent three rounds of subcloning by limiting dilution.
Characterization of MAbs directed against PPV1 VP2 protein
The isotype of produced MAbs was determined using the Mouse MonoAb-ID kit (HRP) (Invitrogen) according to the manufacturer’s instructions. For that purpose, the PPV1-specific IPMA plates were prepared and used according to previous descriptions, where the hybridoma supernatants were added as primary antibodies and monospecific rabbit polyclonal antibodies were added as a secondary antibody. Additionally, a HRP-conjugated goat anti-rabbit antibody was used for the detection of the secondary antibodies. The color reaction was developed by following previous procedures. This test identifies the IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM isotype classes and the κ and λ light chains.
A Western blotting analysis was used to characterize the immunoreactivity of the MAbs with native PPV1 VP2, which was expressed during the infection of ST cells by PPV1. Generally, ST cells (80–90 % confluence) were inoculated with the PPV1-LH strain at a concentration of 105 TCID50/dish and collected at 32 hpi. The cell lysates were separated with 12 % SDS-PAGE and then transferred onto NC membranes. The hybridoma culture supernatants were used as the primary antibodies and an HRP-conjugated rabbit anti-mouse IgG (H + L) antibody (Sigma-Aldrich) was used as the secondary antibody. The SP2/0 culture supernatant was used as the negative antibody control. Antibody binding was evaluated by the addition of the DAB substrate.
Primary scanning of the PPV1 VP2 B cell linear epitopes using VP2-specific MAbs
Three polypeptides (designated A, B, and C) covering the amino acid sequence of PPV1 VP2 were designed and expressed as glutathione S-transferase (GST) fusion proteins. The adjacent polypeptides overlapped by eight amino acids, i.e., polypeptide A corresponded to VP2 amino acids 1–179, polypeptide B to VP2 amino acids 172–372, and polypeptide C to VP2 amino acids 365–579. The primers for the amplification of the three polypeptides are listed in Table S1. The target genes were cloned into the BamHI and XhoI sites of pGEX-6P-1. Escherichia coli BL-21 cells were then transformed with the recombinant plasmids, which were induced by the addition of isopropyl-d-thiogalactopyranoside. A Western blotting analysis was used to evaluate the expression of the recombinant proteins and their reactions with each MAb. A mouse anti-GST-tag MAb (Sigma-Aldrich) and the hybridoma cell culture supernatants were used as the primary antibodies, and an IRDye-700-conjugated goat anti-mouse IgG (H + L) antibody was used as the secondary antibody. The results were detected with an infrared fluorescence scanning imaging system (Licor Odyssey, Lincoln, Nebraska, USA).
Refined scanning of the PPV1 VP2 protein B cell linear epitopes using VP2-specific MAbs
To localize the antibody-binding epitopes of the PPV1 VP2 protein, a panel of 16 peptides was designed (Table 1) and oligonucleotides pairs encoding them (listed in Table S2) were synthesized (BGI, Beijing, China). The oligonucleotide pairs were annealed to each other at 55 °C for 30 min and cloned into the BamHI and XhoI sites of pGEX-6P-1. Escherichia coli BL-21 cells were transformed with the recombinant plasmids to express the recombinant proteins. Sixteen GST-fused peptides were obtained and used as coating antigens, with the GST tag alone used as the negative protein control. Briefly, the bacterial cells were pelleted at 5000×g for 10 min, lysed by sonication in PBS, and then coated onto high-absorption 96-well ELISA plates at 4 °C overnight at a dilution of 1:100 in 50 mM carbonate buffer (pH 9.6). The plates were then washed three times with 50 mM PBS (pH 7.2) containing 0.05 % Tween 20 (PBST), blocked with 1 % BSA–PBST at 37 °C for 1 h, and washed as described above. The hybridoma culture supernatants (100 μL/well) were added and incubated at 37 °C for 1 h, and the plates were washed. Diluted (1:4000) HRP-conjugated rabbit anti-mouse IgG (H + L) antibody (Sigma-Aldrich) was then added and incubated at 37 °C for 1 h. After the plates were washed three times, the colorimetric reaction was developed for 25 min by adding 100 μL of 0.21 mg/mL 2,2-azino-di(3-ethylbenzthiazoline sulfonic acid) in 0.1 M citrate (pH 4.2) containing 0.003 % hydrogen peroxide (ABTS substrate) (Huang et al. 2014). The reactions were stopped by the addition of 50 μL of NaF solution. The iELISA was performed in triplicate and the optical density at 405 nm (OD405) was measured with a microtiter plate reader (Bio-Rad, Hercules, USA).
To define the minimal linear epitopes of PPV1 VP2, another set of 39 complementary oligonucleotide pairs was synthesized (listed in Table S3) and their corresponding peptides were expressed and scanned with an iELISA, as described above. A Western blotting analysis was also conducted to test the immunoreactivity of the PPV1 VP2 core epitopes for their corresponding MAbs. The baculovirus-expressed PPV1 VP2 protein was used as the positive protein control and the GST tag alone as the negative protein control.
Structural analysis of the identified PPV1 VP2 epitopes
The spatial and secondary structures of the three PPV1 VP2 epitopes were analyzed by mapping them onto the virion with a solid surface and onto the VP2 monomer (PDB ID: 1K3V) with or without a solid surface, drawn with the Chimera 1.7.1 software (Pettersen et al. 2004).
Homology analysis of the identified PPV1 VP2 epitopes in different PPV1 strains
To assess the conservation of the three identified linear epitopes of the PPV1 VP2 protein in different PPV1 strains, a collection of 29 PPV1 VP2 amino acid sequences were retrieved from the GenBank database and analyzed with the Quick Alignment method, with a gap penalty of 3, using the DNAMAN v6.0 software (Lynnon BioSoft Inc., San Ramon, CA, USA).
Epitope-based iELISAs
Because epitope 228QQITDA233 is exposed on the virion surface, a VP2-epitope-based iELISAs was established and used to test the anti-PPV1 antibodies produced in immunized animals. The IPMA was used as the reference method and performed as described for screening the hybridoma supernatants. A polypeptide, 216RNLDPPTYTGQSQQITDAIQTGPHSDIMF244, was synthesized (GL Biochem, Shanghai, China) and coated onto high-absorption 96-well ELISA plates at a concentration of 1 μg/well at 4 °C overnight. Serum samples diluted 1:50 with 1 % BSA–PBST were added to the wells, which were then incubated at 37 °C for 1 h. The subsequent procedures were conducted as described for the refined scanning of the PPV1 VP2 epitopes. IPMA-identified PPV1-positive and PPV1-negative guinea pig sera were used as the positive and negative controls, respectively. The optimal conditions for the VP2 epitope-based iELISA were investigated by varying the conditions at each step (except in the colorimetric reaction and reaction termination), while maintaining the conditions at all other steps constant (Huang et al., 2011a, 2011b). The mean (M) and standard deviation (SD) of the OD405 values for 24 negative controls were used to estimate the cutoff level (Hu et al. 2014; Huang et al. 2011a, 2011b). Based on this criterion, a VP2 epitope-based iELISA was established to scan 135 guinea pig sera. The sensitivity, specificity, and concordance rate of the data from the VP2 epitope-based iELISAs relative to those from the IPMA were calculated as previously described (Liu et al. 2004).
MAb-based IFA
ST cells seeded in glass-bottomed cell culture dishes (80–90 % confluence) were inoculated with the PPV1-LH strain at a concentration of 105 TCID50/dish and fixed with prechilled absolute methanol at −20 °C for 10 min at 24, 36, 48, 72, and 96 hpi. After the cells were washed with PBS, they were incubated with MAb 8H6 (diluted 1:1000) and incubated at 37 °C for 1 h. After the cells were washed with PBS, they were incubated with a goat anti-mouse IgG (H + L) secondary antibody (1:800) conjugated with Alexa Fluor® 488 (Life Technologies, Carlsbad, CA, USA) at 37 °C for 30 min. After the cells were washed three times with PBS, the cellular DNA was stained with Hoechst 33258 (Life Technologies) at room temperature for 10 min and washed with PBS. Images were captured with a Leica DM-IRE2 confocal microscope.
Results
Identification of expressed recombinant PPV1-VP2 protein
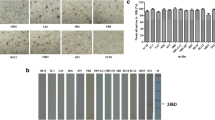
The immunoreactivity of recombinant PPV1 VP2 was identified with a Western blotting analysis. There was a specific band in the recombinant baculovirus-infected Sf21 cell lysates with a molecular weight of about 64 kDa (Fig. 1a). In contrast, no protein was detectable in the normal Sf21 cell lysates or the lysates of Sf21 cells infected with wild-type baculovirus.
a Identification of baculovirus-expressed recombinant PPV1 VP2 protein. Lane M, PageRuler™ Prestained Protein Ladder (Invitrogen, Carlsbad, CA, USA); lane Sf21, cell lysates from normal Sf21 cells; lane VP2, recombinant PPV1 VP2 protein expressed in a baculovirus expression system; and lane Wt, lysate of insect cells infected with wild-type baculovirus. b Scanning monoclonal antibodies (MAbs) with an immunoperoxidase monolayer assay (IPMA). IPMA plates were prepared and used to scan hybridoma supernatants. Three PPV1-specific MAbs (8H6, 4A1, and 8E4) were identified. Normal ST cells were used as the negative control. c Characterization of prepared monoclonal antibodies (MAbs) with Western blotting. The immunoreactivity of native PPV1 VP2 to MAbs was analyzed with Western blotting. The native PPV1 VP2 protein was expressed after PPV1 infection of ST cells. Lane M, PageRuler™ Prestained Protein Ladder (Invitrogen); lane 8H6, MAb 8H6 was used as the primary antibody; lane 4A1, MAb 4A1was used as the primary antibody; lane 8E4, MAb 8E4 was used as the primary antibody; and lane SP2/0, SP2/0 cell supernatant as the negative control
Production and characterization of VP2-specific MAbs
Three hybridoma cell lines that produced stable MAbs against the PPV1 VP2 protein were identified with IPMA (Fig. 1b) and designated 8H6, 4A1, and 8E4. The MAb heavy chain isotype was IgG1 for 8H6 and 8E4, and IgM for 4A1. The MAb light chain isotype was κ-chain for 8H6 and 8E4, and λ for 4A1. All three MAbs reacted well with the native VP2 protein according to a Western blotting analysis (Fig. 1c), but no band was detectable in the SP2/0 negative antibody control.
Primary scanning of PPV1 VP2 B cell linear epitopes using VP2-specific MAbs
The Western blotting analysis indicated that all three GST-fused polypeptides (A, B, and C) were successfully expressed, with protein molecular weights of ∼42.5, ∼44.5, and ∼46 kDa, respectively, and the GST tag alone had a molecular weight of ∼26 kDa (Fig. 2, left). All the MAbs (only one figure is shown) specifically recognized polypeptide B (Fig. 2, right), when the GST tag alone was used as the negative protein control.
Primary scanning of PPV1 VP2 B cell epitopes using VP2-specific monoclonal antibodies (MAbs). Truncated polypeptides A, B, and C were expressed as GST-fused proteins. Polypeptide A corresponded to VP2 amino acids 1–179 with the protein molecular weight of ∼42.5 kDa, polypeptide B corresponded to VP2 amino acids 172–372 with the protein molecular weight of ∼44.5 kDa, polypeptide C corresponded to VP2 amino acid 365–579 with the protein molecular weight of ∼46 kDa. An anti-GST-tag MAb and the prepared VP2-specific MAbs were used separately as the primary antibodies. Lane M, PageRuler™ Prestained Protein Ladder (Invitrogen); lane GST, GST tag alone as the negative control protein; lane A, GST-fused polypeptide A; lane B, GST-fused polypeptide B; lane C, GST-fused polypeptide C
Refined scanning of the B cell linear epitopes of PPV1 VP2 using VP2-specific MAbs
To localize the regions of the PPV1 VP2 protein in which the epitopes occur, 16 peptides (B1–B16) based on the amino acid sequence of polypeptide B were successfully expressed as GST-fused proteins and their reactions with the prepared MAbs were scanned with an iELISA. The results indicated that peptides B5, B10, and B15 were recognized by 8H6, 4A1, and 8E4, respectively (Fig. 3).
Reactivity analysis of peptides B1–B16 with monoclonal antibodies (MAbs) 8H6 (a), 4A1 (b), and 8E4 (c). Peptides B1–B16, based on the amino acid sequence of polypeptide B, were expressed and scanned for their reactivity with MAbs (8H6, 4A1, and 8E4) using indirect enzyme-linked immunosorbent assays. B1 corresponded to VP2 amino acids 172–191, B2 corresponded to VP2 amino acids 184–203, B3 corresponded to VP2 amino acids 196–215, B4 corresponded to VP2 amino acids 208–227, B5 corresponded to VP2 amino acids 220–239, B6 corresponded to VP2 amino acids 232–251, B7 corresponded to VP2 amino acids 244–263, B8 corresponded to VP2 amino acids 256–275, B9 corresponded to VP2 amino acids 268–287, B10 corresponded to VP2 amino acids 280–299, B11 corresponded to VP2 amino acids 292–311, B12 corresponded to VP2 amino acids 304–323, B13 corresponded to VP2 amino acids 316–335, B14 corresponded to VP2 amino acids 328–347, B15 corresponded to VP2 amino acids 340–359, B16 corresponded to VP2 amino acids 352–371. The SP2/0 cell culture supernatant was used as the negative antibody control, and the GST tag with no fused peptide was used as the negative protein control. Error bars indicate the standard deviations of three repeated experiments
Having identified three linear epitopes among these 16 peptides, we progressively truncated peptides B5, B10, and B15 to define the minimal PPV1 VP2 B cell linear epitopes with an iELISA. The removal of residue Q228 at the amino terminus of peptide B5 (220PPTYTGQSQQITDAIQTGPH239) completely reduced the binding by MAb 8H6 (Fig. 4a). Similarly, the deletion of A233 at the carboxyl terminus caused a sharp drop in the OD405 value. This result indicates that both Q228 and A233 are critical residues for the integrity of the epitope, and that 228QQITDA233 (designated B5-E1) is the minimal linear epitope required for optimal 8H6 binding.
Progressively truncated peptides define the PPV1 VP2 core epitopes recognized by MAbs. MAbs 8H6 (a), 4A1 (b), and 8E4 (c) were screened against a series of truncated peptides, with amino acid residues progressively deleted from the amino or carboxyl terminus, to determine the minimal amino acid sequences required for MAb binding. The peptide designations and their corresponding amino acid sequences are given below the figures. The SP2/0 cell culture supernatant was used as the negative antibody control, and the GST tag with no fused peptide was used as the negative protein control. Error bars indicate the standard deviations of three repeated experiments
Further truncation of peptide B10 (280WQTNRSLGLPPKLLTEPTTE299) at residues R284 at the amino terminus and K291 at the carboxyl terminus completely abolished 4A1 binding, demonstrating the importance of these two residues in epitope maintenance (Fig. 4b). This defines 284RSLGLPPK291 (designated B10-E2) as the minimal linear epitope recognized by 4A1.
The peptide B15 (340PYMNFEYSNGGPFLTPIVPT359) was truncated with the same strategy, and F344 and I356 were shown to be important for the constitution of the core epitope recognized by MAb 8E4 (Fig. 4c). The minimal linear epitope was specified as 344FEYSNGGPFLTPI356 (designated B15-E3).
All three PPV1 VP2 core epitopes displayed good immunoreactivity for their corresponding MAbs in a Western blotting analysis (Fig. 5), with protein molecular weight of 26.6, 26.8, and 27.2 kDa, respectively.
Identification of the PPV1 VP2 core epitopes by Western blotting analysis. The immunoreactivity of the three PPV1 VP2 core epitopes with the corresponding MAbs (8H6, 4A1, and 8E4) was confirmed with a Western blotting analysis. Lane M, PageRuler™ Prestained Protein Ladder (Invitrogen); lane VP2, baculovirus-expressed PPV1 VP2 protein as the positive protein control; lane GST, GST tag alone as the negative protein control; lane E1, GST-fused core epitope B5-E1 (228QQITDA233); lane E2, GST-fused core epitope B10-E2 (284RSLGLPP291); and lane E3, GST-fused core epitope B15-E3(344FEYSNGGPFLTPI356)
Structural analysis of the identified PPV1 VP2 core epitopes
The identified PPV1 VP2 core epitopes were mapped onto 3D models of the PPV1 virion and the VP2 monomer. The results showed that only B5-E1 (indicated in red) is exposed on the virion surface, with a random coil secondary structure, whereas B10-E2 (in green) and B15-E3 (in magenta) are located inside the virion capsid protein, with β-turn and β-pleated sheet secondary structures, respectively (Fig. 6).
Mapping the locations of the identified PPV1 VP2 core epitopes on 3D models of the virion and VP2 monomer. a Spatial distributions of the three core epitopes in the complete PPV1 virion with a solid surface (228QQITDA233 is indicated in red, 284RSLGLPPK291 in green, and 344FEYSNGGPFLTPI356 in magenta); b secondary structural analysis of the three core epitopes on the PPV1 VP2 monomer skeleton; c stereostructures of the three core epitopes in the PPV1 VP2 protein monomer with a solid surface. Cartoons of the complete PPV1 virions and PPV1 VP2 monomer were drawn with the Chimera 1.7.1 software, and the identified core epitopes were mapped onto the constructed stereostructures
Homology analysis of the identified PPV1 VP2 epitopes in different PPV1 strains
An alignment of 29 amino acid sequences of PPV1 VP2 proteins showed that B5-E1 (228QQITDA233) is highly conserved, because in only seven of the 29 VP2 sequences was the serine at residue 233 substituted with threonine or alanine. However, B10-E2 (284RSLGLPPK291) and B15-E3 (344FEYSNGGPFLTPI356) are completely conserved, with no amino acid change in any of the sequences (Table 2).
Evaluation of VP2 epitope-based iELISA for the serodiagnosis of anti-PPV1 antibodies
To determine the cutoff value for the VP2-epitope-based iELISAs, 24 negative serum samples (determined with IPMA) were included and analyzed. The cutoff value was defined as an OD405 of 0.24 (M + 3SD). In comparative experiments with IPMA using 135 guinea pig sera, the VP2 epitope-based iELISA achieved a sensitivity of 83.33 %, a specificity of 85.47 %, and a coincidence rate of 85.19 % with the data obtained with the IPMA (Table 3).
MAb-based IFA to characterize PPV1 replication
MAb 8H6 was used to establish an IFA with which to monitor of the PPV1 VP2 protein during viral replication. PPV1 VP2 was detected around the nucleus at 24 hpi, whereas the protein tended to polarize towards the cellular membrane from 36 to 72 hpi (Fig. 7). The dynamic state of the PPV1 VP2 protein indicates that the newly encapsulated virions were released from the nucleus at 24 hpi, so that the replication cycle of PPV1 took less than 24 h.
Discussion
PPV1 is a causative agent of reproductive failure in swine, posing a great economic threat to the farming industry and international trade (Kim and Chae 2004). Recently, a high mutation rate, similar to that of the RNA viruses, was reported in PPV1, with an evolutionary rate of approximately 10−4 substitutions per site per year for the capsid genes (Streck et al. 2011). It is suggested that certain domains have lower evolutionary rates as a consequence of functional constraints. In contrast, dominant mutations have evolved more rapidly than synonymous substitutions in response to advantageous selection (Bergeron et al. 1993; Cadar et al. 2013; Lukashov and Goudsmit 2001; Martins Soares et al. 2003; Streck et al. 2011; Zimmermann et al. 2006). These different evolutionary rates may indicate that the virus uses antigenic shift to resist selective pressures without disturbing the integrity of certain functions. Because vaccine development is always strongly associated with viral evolution, changes in viral immunogenicity during the antigenic evolution of PPV1 might influence the updating of vaccines. Here, we focused on the immunodominant protein of PPV1, the VP2 protein, as a potential candidate for development of diagnostic and research reagents. Analysis of the PPV1 VP2 epitopes should extend our understanding of PPV1-specific immunity and accelerate the development of epitope-based diagnostic tools and synthesized peptide vaccine for PPV1 (Hu et al. 2014; Kamstrup et al. 1998). The report of Kamstrup et al. (1998) firstly described a total of nine linear epitopes of both PPV1 VP1 and VP2 proteins and their immunogenicity was subsequently investigated. The results indicated that, among all the identified antigenic sites, only peptides from the N-terminal part of VP2 were able to induce virus-neutralizing antibodies, although at low levels (Kamstrup et al. 1998). The crystal structure of PPV1 capsids expressed by a baculovirus-based system was initially solved using X-ray crystallography by Simpson et al. (2002), and the degree of conservation of surface-exposed residues was found to be lower than that of canine parvovirus and minute virus of mice, suggesting potential variability in antigenicity and host range (Simpson et al. 2002).
To better understand the antigenic structure of PPV1 VP2 protein, we developed three PPV1 VP2-specific MAbs and used them to map the VP2 protein B cell linear epitopes in this study. Recombinant PPV1 VP2 protein was successfully expressed by the baculovirus expression system to recapitulate its natural morphology and used for immunization of mice (Antonis et al. 2006; Qi and Cui, 2009; Wang et al. 2015). The IPMA was applied for screening of hybridoma cell supernatants to exclude potential false positive results for VP2-reactive MAbs production. A PEPSCAN analysis of a panel of 58 GST-fused proteins identified 228QQITDA233 (B5-E1), 284RSLGLPP291 (B10-E2), and 344FEYSNGGPFLTPI356 (B15-E3) as the three VP2 core epitopes using MAbs against recombinant VP2 protein. Of the three epitopes identified, B10-E2 and B15-E3 are reported for the first time in this study, whereas B5-E1 was included in the earlier report (Kamstrup et al. 1998).
A structural analysis indicated that epitope B5-E1 is exposed on the virion surface, whereas B10-E2 and B15-E3 are located inside the virion. Their secondary structures indicate that B5-E1 is more flexible, with random coiling, whereas B15-E3 shows strong conformational stability because it forms a β-pleated sheet, and B10-E2 forms a β-turn structure arising from the two consecutive prolines in its sequence. This is the first illustration of the VP2 epitopes in 3D models. MAb 8H6 might be applicable to the development of other diagnostic methods, such as an antigen capture ELISA, because its epitope is exposed on the virion surface.
An alignment of the amino acid sequences of various PPV1 VP2 proteins indicated that B10-E2 and B15-E3 are completely conserved among the 29 tested PPV1 strains from different regions, whereas B5-E1 contains a single amino acid substitution at residue 233. This suggests that the epitope exposed on the virion surface, with a flexible secondary structure, is more likely to mutate than those located inside the virion, with stable conformations. The PPV1 VP2 core epitopes identified here could provide important insights into the structure and function of the VP2 and may play important roles in the antigenic evolution of PPV1 (Cadar et al. 2013).
Generally, antigenic sites located outside the complete virion are more accessible for antibody binding. Because epitope B5-E1 is exposed on the virion surface, a VP2 epitope-based iELISA was preliminarily established and evaluated for the assay of anti-PPV1 antibodies in the sera of guinea pigs. Because of the limitations entailed in using a single epitope, the coincidence rate of the established VP2 epitope-based iELISA (85.19 %) in a comparative analysis with IPMA might not seem very satisfactory. However, the iELISA was more rapid and the interpretation of the results was more objective, with a higher throughput than was possible with IFA or IPMA. Epitope-based diagnostic methods could effectively avoid the proliferation of infectious viruses, suggesting that the use of iELISA for diagnostic purposes should be investigated further.
The cellular locations of the VP2 protein at different times during viral infection were monitored with confocal microscopy. The localization of PPV1 is reported to be perinuclear within 4 hpi, and new virus was observed in the nucleus at 16–20 hpi (Boisvert et al. 2010). In this study, we confirmed that newly synthesized virions are released from the nucleus at 24 hpi, so the viral replication cycle takes less than 24 h. MAb-based IFA may provide some tools for monitoring viral antigens in vitro and for pathway analyses during PPV1 infection.
In conclusion, we prepared three PPV1 VP2-specific MAbs, defined their antigenic core epitopes, and demonstrated the potential application of VP2 epitope-based iELISA to the serodiagnosis of anti-PPV1 antibodies in guinea pig sera and a MAb-based IFA for the detection of viral antigens. To the best of our knowledge, this is the first time that the linear core epitopes of the PPV1 VP2 protein have been defined using MAbs and a serodiagnosis method established based on the identified epitopes of PPV1 VP2. The three B cell linear epitopes of PPV1 VP2 described here should allow us to clarify the molecular mechanisms of antigenic evolution.
References
Adlhoch C, Kaiser M, Ellerbrok H, Pauli G (2010) High prevalence of porcine hokovirus in German wild boar populations. Virol J 7:171
Antonis AF, Bruschke CJ, Rueda P, Maranga L, Casal JI, Vela C, Hilgers LA, Belt PB, Weerdmeester K, Carrondo MJ, Langeveld JP (2006) A novel recombinant virus-like particle vaccine for prevention of porcine parvovirus-induced reproductive failure. Vaccine 24(26):5481–5490
Bergeron J, Menezes J, Tijssen P (1993) Genomic organization and mapping of transcription and translation products of the NADL−2 strain of porcine parvovirus. Virology 197(1):86–98
Boisvert M, Fernandes S, Tijssen P (2010) Multiple pathways involved in porcine parvovirus cellular entry and trafficking toward the nucleus. J Virol 84(15):7782–7792
Boisvert M, Bouchard-Levesque V, Fernandes S, Tijssen P (2014) Classic nuclear localization signals and a novel nuclear localization motif are required for nuclear transport of porcine parvovirus capsid proteins. J Virol 88(20):11748–11759
Cadar D, Csagola A, Kiss T, Tuboly T (2013) Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol Phylogenet Evol 66(1):243–253
Cheung AK, Wu G, Wang D, Bayles DO, Lager KM, Vincent AL (2010) Identification and molecular cloning of a novel porcine parvovirus. Arch Virol 155(5):801–806
Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ (2014) The family Parvoviridae. Arch Virol 159(5):1239–1247
Fernandes S, Boisvert M, Tijssen P (2011) Genetic elements in the VP region of porcine parvovirus are critical to replication efficiency in cell culture. J Virol 85(6):3025–3029
Hijikata M, Abe K, Win KM, Shimizu YK, Keicho N, Yoshikura H (2001) Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn J Infect Dis 54(6):244–245
Hu Z, Chang H, Ge M, Lin Y, Wang X, Guo W, Wang X (2014) Development of antigen capture ELISA for the quantification of EIAV p26 protein. Appl Microbiol Biotechno l 98(21):9073–9081
Huang L, Lu Y, Wei Y, Guo L, Liu C (2011a) Development of a blocking ELISA for detection of serum neutralizing antibodies against porcine circovirus type 2. J Virol Methods 171(1):26–33
Huang L, Lu Y, Wei Y, Guo L, Wu H, Zhang F, Fu Y, Liu C (2011b) Construction and biological characterisation of recombinant porcine circovirus type 2 expressing the V5 epitope tag. Virus Res 161(2):115–123
Huang L, Zhang F, Tang Q, Wei Y, Wu H, Guo L, Fu Y, Liu C (2014) A recombinant porcine circovirus type 2 expressing the VP1 epitope of the type O foot-and-mouth disease virus is infectious and induce both PCV2 and VP1 epitope antibodies. Appl Microbiol Biotechnol 98(22):9339–9350
Kamstrup S, Langeveld J, Botner A, Nielsen J, Schaaper WM, Boshuizen RS, Casal JI, Hojrup P, Vela C, Meloen R, Dalsgaard K (1998) Mapping the antigenic structure of porcine parvovirus at the level of peptides. Virus Res 53(2):163–173
Kim J, Chae C (2004) Concurrent presence of porcine circovirus type 2 and porcine parvovirus in retrospective cases of exudative epidermitis in pigs. Vet J 167(1):104–106
Liu CM, Ihara T, Nunoya T, Ueda S (2004) Development of an ELISA based on the baculovirus-expressed capsid protein of porcine circovirus type 2 as antigen. J Vet Med Sci 66(3):237–242
Lukashov VV, Goudsmit J (2001) Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J Virol 75(6):2729–2740
Mahnel H, Mayr A, Bibrack B (1966) Multiplication of swine fever virus with cytopathogenic effect in piglet testis culture. Zentralbl Veterinarmed B 13(3):250–259
Maranga L, Brazao TF, Carrondo MJ (2003) Virus-like particle production at low multiplicities of infection with the baculovirus insect cell system. Biotechnol Bioeng 84(2):245–253
Martins Soares R, Cortez A, Heinemann MB, Sakamoto SM, Martins VG, Bacci M, De Campos Fernandes FM, Richtzenhain LJ (2003) Genetic variability of porcine parvovirus isolates revealed by analysis of partial sequences of the structural coding gene VP2. J Gen Virol 84(Pt 6):1505–1515
Ni J, Qiao C, Han X, Han T, Kang W, Zi Z, Cao Z, Zhai X, Cai X (2014) Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol J 11(1):203
Nuesch JP, Rommelaere J (2006) NS1 interaction with CKII alpha: novel protein complex mediating parvovirus-induced cytotoxicity. J Virol 80(10):4729–4739
Nuesch JP, Dettwiler S, Corbau R, Rommelaere J (1998) Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J Virol 72(12):9966–9977
Opriessnig T, Fenaux M, Yu S, Evans RB, Cavanaugh D, Gallup JM, Pallares FJ, Thacker EL, Lager KM, Meng XJ, Halbur PG (2004) Effect of porcine parvovirus vaccination on the development of PMWS in segregated early weaned pigs coinfected with type 2 porcine circovirus and porcine parvovirus. Vet Microbio l 98(3–4):209–220
Opriessnig T, Xiao CT, Gerber PF, Halbur PG (2014) Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet Microbiol 173(1–2):9–16
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Qi T, Cui S (2009) Expression of porcine parvovirus VP2 gene requires codon optimized E. coli cells. Virus Genes 39(2):217–222
Rhode SL (1989) Both excision and replication of cloned autonomous parvovirus DNA require the NS1 (rep) protein. J Virol 63(10):4249–4256
Rueda P, Fominaya J, Langeveld JP, Bruschke C, Vela C, Casal JI (2000) Effect of different baculovirus inactivation procedures on the integrity and immunogenicity of porcine parvovirus-like particles. Vaccine 19(7–8):726–734
Simpson AA, Hebert B, Sullivan GM, Parrish CR, Zadori Z, Tijssen P, Rossmann MG (2002) The structure of porcine parvovirus: comparison with related viruses. J Mol Biol 315(5):1189–1198
Streck AF, Bonatto SL, Homeier T, Souza CK, Goncalves KR, Gava D, Canal CW, Truyen U (2011) High rate of viral evolution in the capsid protein of porcine parvovirus. J Gen Virol 92(Pt 11):2628–2636
Sun J, Huang L, Wei Y, Wang Y, Chen D, Du W, Wu H, Liu C (2015) Prevalence of emerging porcine parvoviruses and their co-infections with porcine circovirus type 2 in China. Arch Virol 160(5):1339–1344
Wang F, Wei Y, Zhu C, Huang X, Xu Y, Yu L, Yu X (2010) Novel parvovirus sublineage in the family of Parvoviridae. Virus Genes 41(2):305–308
Wang YP, Liu D, Guo LJ, Tang QH, Wei YW, Wu HL, Liu JB, Li SB, Huang LP, Liu CM (2013) Enhanced protective immune response to PCV2 subunit vaccine by co-administration of recombinant porcine IFN-gamma in mice. Vaccine 31(5):833–838
Wang HX, Sun EC, Xu QY, Yang T, Zhang Q, Feng YF, Li JP, Lv S, Sun L, Sun J, Wu DL (2015) Analysis of murine B-cell epitopes on bluetongue virus 12 nonstructural protein 1. Appl Microbiol Biotechnol 99(3):1309–1321
Xie HL, Wang Z, Cui SJ, Zhang CF, Cui YD (2010) The epitope of the VP1 protein of porcine parvovirus. Virol J 7:161
Xu Y, Li Y (2007) Induction of immune responses in mice after intragastric administration of Lactobacillus casei producing porcine parvovirus VP2 protein. Appl Environ Microbiol 73(21):7041–7047
Zimmermann P, Ritzmann M, Selbitz HJ, Heinritzi K, Truyen U (2006) VP1 sequences of German porcine parvovirus isolates define two genetic lineages. J Gen Virol 87(Pt 2):295–301
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 31302110), the Public Welfare Special Funds for Agricultural Scientific Research (No. 201203039) and the National High Technology R&D program (863) of China (No. 2011AA10A208).
Conflict of interest
The authors declare that they have no competing interests.
Compliance with ethical standards
Animal welfare and experimental procedures were carried out strictly according to the American Physiological Society’s Guiding Principles for the Care and Use of Animals and approved by Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The animal ethics committee approval number is Heilongjiang-SYXK-2006-032.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jianhui Sun and Liping Huang contributed equally to this article.
Electronic supplementary material
ESM 1
(PDF 108 kb)
Rights and permissions
About this article
Cite this article
Sun, J., Huang, L., Wei, Y. et al. Identification of three PPV1 VP2 protein-specific B cell linear epitopes using monoclonal antibodies against baculovirus-expressed recombinant VP2 protein. Appl Microbiol Biotechnol 99, 9025–9036 (2015). https://doi.org/10.1007/s00253-015-6790-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6790-z