Abstract
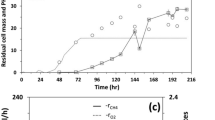
Long chain fatty acids (LCFAs) are important intermediates in the anaerobic degradation of n-alkanes. In order to find out the biochemical processes involved in the degradation of LCFAs, palmitate (a typical LCFA) was used as a substrate, and low-temperature oilfield production fluids were used as a source of microorganisms to establish two anaerobic systems, one with addition of sulfate as exogenous electron acceptor (SP), another without exogenous electron acceptor (MP) and both incubated at room temperature. After more than 2 years of incubation, about 48 and 57.4 % of the palmitate were degraded in samples of MP and SP, respectively. Methane production reached 1408 and 1064 μmol for MP and SP, respectively. Clone libraries of archaeal 16S rRNA genes showed that the predominant archaea in the sulfate-amended cultures (SP) was Methanosaeta whereas Methanocalculus dominated the culture without addition of exogenous sulfate (MP). This observation shows that palmitate could be biodegraded into methane through β-oxidation and acetoclastic methanogenesis in the presence of with or without sulfate. The high occurrence of Methanosaeta in the sulfate-amended system indicates that acetoclastic methanogenesis was not inhibited/little affected by the addition of sulfate. Acetoclastic methanogenesis might be the predominant biochemchimcal pathway of methane generation in enrichment cultures amended with sulfate. These results shed light on alternative methanogenic pathways in the presence of sulfate.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Amend JP, Shock EL (2001) Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev 25:175–243. doi:10.1111/j.1574-6976.2001.tb00576.x
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741. doi:10.1128/AEM.00556-06
Beckmann S, Lueders T, Krüger M, Von Netzer F, Engelen B, Cypionka H (2011) Acetogens and acetoclastic Methanosarcinales govern methane formation in abandoned coal mines. Appl Environ Microbiol 77:3749–3756. doi:10.1128/aem.02818-10
Callaghan AV, Gieg LM, Kropp KG, Suflita JM, Young LY (2006) Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium. Appl Environ Microbiol 72:4274–4282. doi:10.1128/aem.02896-05
Callbeck CM, Agrawal A, Voordouw G (2013) Acetate production from oil under sulfate-reducing conditions in bioreactors injected with sulfate and nitrate. Appl Environ Microbiol 79:5059–5068. doi:10.1128/aem.01251-13
Cheng L, Qiu TL, Yin XB, Wu XL, Hu GQ, Deng Y, Zhang H (2007) Methermicoccus shengliensis gen. nov., sp. nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov. Int J Syst Evol Microbiol 57:2964–2969. doi:10.1099/ijs.0.65049-0
Cheng L, Shi S, Li Q, Chen J, Zhang H, Lu Y (2014) Progressive degradation of crude oil n-alkanes coupled to methane production under mesophilic and thermophilic conditions. PLoS ONE 9, e113253. doi:10.1371/journal.pone.0113253
Colleran E, Finnegan S, Lens P (1995) Anaerobic treatment of sulphate-containing waste streams. Antonie Leeuwenhoek 67:29–46. doi:10.1007/bf00872194
Cravo-Laureau C, Grossi V, Raphel D, Matheron R, Hirschler-Réa A (2005) Anaerobic n-alkane metabolism by a sulfate-reducing bacterium, Desulfatibacillum aliphaticivorans Strain CV2803T. Appl Environ Microbiol 71:3458–3467. doi:10.1128/aem.71.7.3458-3467.2005
Dolfing J (2014) Thermodynamic constraints on syntrophic acetate oxidation. Appl Environ Microbiol 80:1539–1541. doi:10.1128/aem.03312-13
Dolfing J, Larter SR, Head IM (2008) Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J 2:442–452. doi:10.1038/ismej.2007.111
Dolfing J, Xu A, Gray ND, Larter SR, Head IM (2009) The thermodynamic landscape of methanogenic PAH degradation. Microb Biotechnol 2:566–574. doi:10.1111/j.1751-7915.2009.00096.x
Embree M, Nagarajan H, Movahedi N, Chitsaz H, Zengler K (2014) Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J 8:757–767. doi:10.1038/ismej.2013.187
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi:10.1093/bioinformatics/bts565
Grabowski A, Blanchet D, Jeanthon C (2005) Characterization of long-chain fatty-acid-degrading syntrophic associations from a biodegraded oil reservoir. Res Microbiol 156:814–821. doi:10.1016/j.resmic.2005.03.009
Gray ND, Sherry A, Grant RJ, Rowan AK, Hubert CRJ, Callbeck CM, Aitken CM, Jones DM, Adams JJ, Larter SR, Head IM (2011) The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol 13:2957–2975. doi:10.1111/j.1462-2920.2011.02570.x
Guan J, Xia LP, Wang LY, Liu JF, Gu JD, Mu BZ (2013) Diversity and distribution of sulfate-reducing bacteria in four petroleum reservoirs detected by using 16S rRNA and dsrAB genes. Int Biodeterior Biodegrad 76:58–66. doi:10.1016/j.ibiod.2012.06.021
Guan J, Zhang BL, Mbadinga S, Liu JF, Gu JD, Mu BZ (2014) Functional genes (dsr) approach reveals similar sulphidogenic prokaryotes diversity but different structure in saline waters from corroding high temperature petroleum reservoirs. Appl Environ Microbiol 98:1871–1882. doi:10.1007/s00253-013-5152-y
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319. doi:10.1093/bioinformatics/bth226
Jones DM, Watson JS, Meredith W, Chen M, Bennett B (2000) Determination of naphthenic acids in crude oils using nonaqueous ion exchange solid-phase extraction. Anal Chem 73:703–707. doi:10.1021/ac000621a
Kendall M, Boone D (2006) The Order Methanosarcinales in Dworkin M, Falkow S, Rosenberg E, Schleifer K-H and Stackebrandt E (eds) The Prokaryotes, Springer, New York, pp 244-256. doi: 10.1007/0-387-30743-5_12
Kuever J (2014a) The Family Desulfarculaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson EF (eds) The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Springer, Berlin, pp 41–44. doi:10.1007/978-3-642-39044-9_270
Kuever J (2014b) The Family Desulfobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson EF (eds) The Prokaryotes. Springer, Berlin, pp 45–73. doi:10.1007/978-3-642-39044-9_266
Kuever J (2014c) The Family Desulfobulbaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson EF (eds) The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Springer, Berlin, pp 75–86. doi:10.1007/978-3-642-39044-9_267
Lai MC, Chen SC, Shu CM, Chiou MS, Wang CC, Chuang MJ, Hong TY, Liu CC, Lai LJ, Hua JJ (2002) Methanocalculus taiwanensis sp. nov., isolated from an estuarine environment. Int J Syst Evol Microbiol 52:1799–1806. doi:10.1099/ijs.0.01730-0
Lee ZM, Bussema C 3rd, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37(database issue):D489–D493. doi:10.1093/nar/gkn689
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics/Comput Appl Biosci 22:1658–1659. doi:10.1093/bioinformatics/btl158
Mayumi D, Dolfing J, Sakata S, Maeda H, Miyagawa Y, Ikarashi M, Tamaki H, Takeuchi M, Nakatsu CH, Kamagata Y (2013) Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat Commun 4.: 1998 doi:10.1038/ncomms2998
Mbadinga SM, Wang LY, Zhou L, Liu JF, Gu JD, Mu BZ (2011) Microbial communities involved in anaerobic degradation of alkanes. Int Biodeterior Biodegrad 65:1–13. doi:10.1016/j.ibiod.2010.11.009
Mbadinga S, Li KP, Zhou L, Wang LY, Yang SZ, Liu JF, Gu JD, Mu BZ (2012) Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl Environ Microbiol 96:531–542. doi:10.1007/s00253-011-3828-8
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25. doi:10.1093/nar/gkh435
Meredith W, Kelland SJ, Jones DM (2000) Influence of biodegradation on crude oil acidity and carboxylic acid composition. Org Geochem 31:1059–1073. doi:10.1016/S0146-6380(00)00136-4
Noor E, Haraldsdóttir HS, Milo R, Fleming RM (2013) Consistent estimation of Gibbs energy using component contributions. PLoS Comput Biol 9, e1003098. doi:10.1371/journal.pcbi.1003098
Pham VD, Hnatow LL, Zhang S, Fallon RD, Jackson SC, Tomb JF, Delong EF, Keeler SJ (2009) Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ Microbiol 11:176–187. doi:10.1111/j.1462-2920.2008.01751.x
Qu X, Vavilin VA, Mazéas L, Lemunier M, Duquennoi C, He PJ, Bouchez T (2009) Anaerobic biodegradation of cellulosic material: Batch experiments and modelling based on isotopic data and focusing on aceticlastic and non-aceticlastic methanogenesis. Waste Manag 29:1828–1837. doi:10.1016/j.wasman.2008.12.008
Rabus R, Hansen T, Widdel F (2013) Dissimilatory sulfate- and sulfur-reducing Prokaryotes. In: Rosenberg E, Delong E, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin, pp 309–404. doi:10.1007/978-3-642-30141-4_70
Savage KN, Krumholz LR, Gieg LM, Parisi VA, Suflita JM, Allen J, Philp RP, Elshahed MS (2010) Biodegradation of low-molecular-weight alkanes under mesophilic, sulfate-reducing conditions: metabolic intermediates and community patterns. FEMS Microbiol Ecol 72:485–495. doi:10.1111/j.1574-6941.2010.00866.x
Shimizu S, Upadhye R, Ishijima Y, Naganuma T (2011) Methanosarcina horonobensis sp. nov., a methanogenic archaeon isolated from a deep subsurface miocene formation. Int J Syst Evol Micr 61:2503–2507. doi:10.1099/ijs.0.028548-0
Smith KS, Ingram-Smith C (2007) Methanosaeta, the forgotten methanogen? Trends Microbiol 15:150–155. doi:10.1016/j.tim.2007.02.002
Sousa DZ, Pereira MA, Stams AJ, Alves MM, Smidt H (2007) Microbial communities involved in anaerobic degradation of unsaturated or saturated long-chain fatty acids. Appl Environ Microbiol 73:1054–1064. doi:10.1128/AEM.01723-06
Sousa DZ, Alves JI, Alves MM, Smidt H, Stams AJ (2009) Effect of sulfate on methanogenic communities that degrade unsaturated and saturated long‐chain fatty acids (LCFA). Environ Microbiol 11:68–80. doi:10.1111/j.1462-2920.2008.01740.x
Sousa DZ, Balk M, Alves M, Schink B, Mcinerney MJ, Smidt H, Plugge CM, Stams AJM (2010) Degradation of long-chain fatty acids by sulfate-reducing and methanogenic communities. In: Timmis K (ed) Handbook of Hydrocarbon and Lipid Microbiology, 1st edn. Springer, Berlin, pp 963–980. doi:10.1007/978-3-540-77587-4_69
Tan B, Nesbo C, Foght J (2014) Re-analysis of omics data indicates Smithella may degrade alkanes by addition to fumarate under methanogenic conditions. ISME J 8:2353–2356. doi:10.1038/ismej.2014.87
Thauer RK, Jungermann K, Decker K (1977) Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41(1):100–180
Wang LY, Gao CX, Mbadinga SM, Zhou L, Liu JF, Gu JD, Mu BZ (2011) Characterization of an alkane-degrading methanogenic enrichment culture from production water of an oil reservoir after 274 days of incubation. Int Biodeterior Biodegrad 65:444–450. doi:10.1016/j.ibiod.2010.12.010
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679. doi:10.1002/bit.20347
Zellner G, Messner P, Winter J, Stackebrandt E (1998) Methanoculleus palmolei sp. nov., an irregularly coccoid methanogen from an anaerobic digester treating wastewater of a palm oil plant in North-Sumatra, Indonesia. Int J Syst Bacteriol 48:1111–1117. doi:10.1099/00207713-48-4-1111
Zhou L, Li KP, Mbadinga S, Yang SZ, Gu JD, Mu BZ (2012) Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology 21:1680–1691. doi:10.1007/s10646-012-0949-5
Zhu J, Zheng H, Ai G, Zhang G, Liu D, Liu X, Dong X (2012) The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS ONE 7, e36756. doi:10.1371/journal.pone.0036756
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 51174092, 41373070, 31200101), the National Natural Science Foundation of China, and the Research Grants Council Joint Research Fund (Grant No. 41161160560).
Conflict of interest
The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 230 kb)
Rights and permissions
About this article
Cite this article
Lv, L., Mbadinga, S.M., Wang, LY. et al. Acetoclastic methanogenesis is likely the dominant biochemical pathway of palmitate degradation in the presence of sulfate. Appl Microbiol Biotechnol 99, 7757–7769 (2015). https://doi.org/10.1007/s00253-015-6669-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6669-z