Abstract
In comparison to humans and chimpanzees, gorillas show low diversity at MHC class I genes (Gogo), as reflected by an overall reduced level of allelic variation as well as the absence of a functionally important sequence motif that interacts with killer cell immunoglobulin-like receptors (KIR). Here, we use recently generated large-scale genomic sequence data for a reassessment of allelic diversity at Gogo-C, the gorilla orthologue of HLA-C. Through the combination of long-range amplifications and long-read sequencing technology, we obtained, among the 35 gorillas reanalyzed, three novel full-length genomic sequences including a coding region sequence that has not been previously described. The newly identified Gogo-C*03:01 allele has a divergent recombinant structure that sets it apart from other Gogo-C alleles. Domain-by-domain phylogenetic analysis shows that Gogo-C*03:01 has segments in common with Gogo-B*07, the additional B-like gene that is present on some gorilla MHC haplotypes. Identified in ~ 50% of the gorillas analyzed, the Gogo-C*03:01 allele exclusively encodes the C1 epitope among Gogo-C allotypes, indicating its important function in controlling natural killer cell (NK cell) responses via KIR. We further explored the hypothesis whether gorillas experienced a selective sweep which may have resulted in a general reduction of the gorilla MHC class I repertoire. Our results provide little support for a selective sweep but rather suggest that the overall low Gogo class I diversity can be best explained by drastic demographic changes gorillas experienced in the ancient and recent past.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Molecules encoded by the MHC class I genes are an integral component of the adaptive and innate immune system. Their primary function is to bind antigens of intracellular origin and present them on the cell surface to immunocompetent cells. Upon recognition of pathogen-derived antigens, cytotoxic CD8+ T cells are activated to initiate an appropriate immune response, ultimately leading to the lysis of the infected cell (Davis and Bjorkman 1988; Wong and Pamer 2003). Some MHC class I allotypes also interact with NK cells via KIR. Cells showing reduced or aberrant expression of MHC class I molecules due to viral infection or malignant transformation are eliminated by NK cells whose cytolytic activity is regulated by activating and inhibitory KIR (Vilches and Parham 2002; Lanier 2005).
During primate evolution, the MHC class I multigene family has recurrently expanded and contracted in structure and gene number (Kulski et al. 2002; Kelley et al. 2005; Bontrop 2006). As a result of the “birth-and-death” evolutionary process, the MHC class I multigene family consists of a variety of genes: highly polymorphic genes encoding classical MHC class I molecules, conserved genes encoding nonclassical MHC class I molecules, and various pseudogenes and gene fragments (Piontkivska and Nei 2003; Nei and Rooney 2005). Further, the varied coevolutionary dynamics between pathogens and their hosts have shaped the genes encoding the MHC class I molecules leading to species-specific patterns of intra- and interlocus variability (Hughes and Nei 1989; Adams and Parham 2001).
Indeed, orthologues of all classical human MHC class I genes (HLA-A, -B, and -C) can be found in the MHC of the gorilla (Gogo-A, -B, and -C), but not vice versa. Distinguishing the gorilla MHC is a polymorphic A-related locus present on some haplotypes, designated Gogo-Oko, which has no functional human orthologue (Lawlor et al. 1991; Watkins et al. 1991; Gleimer et al. 2011; Hans et al. 2017). Furthermore, there is evidence for differences in the copy number of certain Gogo class I genes. Whereas all human haplotypes have a single copy of the classical HLA class I genes, some gorilla MHC haplotypes are characterized by an additional B-like gene (Hans et al. 2017). However, despite gorillas having a comparatively complex haplotype structure, their MHC class I diversity appears to be low, which is reflected not only by an overall reduced level of allelic variation but also by the absence of a functionally important sequence motif at Gogo-C, the gorilla orthologue of HLA-C (Hans et al. 2017). Specifically, all previously described Gogo-C allotypes are predicted to express only one of two KIR-binding epitopes, the C2 epitope, whereas the C1 epitope is carried only by a minority of Gogo-B allotypes (Lawlor et al. 1991; Urvater et al. 2001; Adams and Parham 2001; Martínez-Laso et al. 2006; Hans et al. 2017). In contrast, MHC-C allotypes of humans and chimpanzees carry either the C1 or the C2 epitope, which suggests that gorillas have lost the C1-bearing allotypes at this locus (Moesta et al. 2009; Parham et al. 2012; Hans et al. 2017).
Here, we extend our previous work characterizing MHC class I genes in gorillas. By using recently generated long-read genomic sequence data, we provide a reassessment of the Gogo-C gene variation, revealing that gorillas possess a single Gogo-C allele that expresses the C1 ligand for KIR. In addition, we extend analyses exploring the hypothesis whether gorillas experienced a selective sweep which might have resulted in the reduction of their MHC class I repertoire.
Material and methods
Virtual MHC genotyping and primer design
Recently, a high-quality genome assembly of the female western lowland gorilla (Gorilla gorilla gorilla) named “Susie” was generated using long-read sequencing technology (Gordon et al. 2016). To investigate the Gogo class I alleles of “Susie,” we downloaded from the European Nucleotide Archive (ENA) the contig (000730_F) spanning the MHC class I region. Comparison to our previously reported full-length Gogo class I alleles (Hans et al. 2017) showed that this haplotype contains the following alleles: Gogo-A*01:01:01, Gogo-A*05:02:01:02N, and Gogo-B*01:01:01:01. However, in the expected genomic position of Gogo-C located approximately 85 kb upstream of Gogo-B, the contig showed only partial sequence similarity to known Gogo-C alleles. Virtual genotyping with the Gogo-C primers used in our previous study revealed that the contig sequence contains a single base pair insertion causing mismatches at the 3′-end of the reverse primer located in the 3′ untranslated region (3’UTR) (Online Resource 1). Although it must be noted that PacBio sequencing errors predominantly consist of single insertions and deletions, these findings strongly suggest that we may have underestimated allelic variation at the Gogo-C gene in our previous study due to the non-amplification of certain alleles (Ross et al. 2013; Laehnemann et al. 2016; Hans et al. 2017).
To assess this possibility, we designed a new primer pair for the long-range PCR (LR-PCR) amplification of Gogo-C to re-evaluate allelic diversity at this locus. From the abovementioned contig, we extracted the sequence comprising the putative Gogo-C gene and generated a ClustalW multiple alignment together with all available MHC class I gene sequences from human, chimpanzee, bonobo, gorilla, and orangutan using BioEdit version 7.2.0 (Hall 1999). To avoid co-amplification of Gogo-B alleles, we also designed a new forward primer. Encompassing complete coding region sequences, primers were manually placed in interspecies conserved regions unique to MHC-C with the forward primer (5′-ACTCCCATTGGGTGTCGGGTTCTAG-3′) located in the enhancer-promoter region and the reverse primer (5′-GGYGTGAAGAAATCCTGCATCTCAGTC-3′) located in the 3’UTR. Primers were designed to have melting temperatures of at least 64 °C to enhance binding specificity (Hans et al. 2017).
Samples, LR-PCR amplifications, and sequencing
High-quality genomic DNAs extracted from 35 gorilla samples were used for the LR-PCR amplifications. These included one eastern lowland gorilla (Gorilla beringei graueri), one Cross River gorilla (Gorilla gorilla diehli), and 33 western lowland gorillas (Gorilla gorilla gorilla). Details of samples and DNA extractions are given in Hans et al. (2017). Two separate LR-PCR amplifications were performed on each DNA sample. In a total volume of 50 μL, LR-PCR reactions contained 1× Crimson LongAmp Taq Reaction Buffer (New England Biolabs, Frankfurt am Main, Germany), 25 μM each dNTP, 0.2 μM each forward and reverse primers, 0.1 U Crimson LongAmp Taq DNA Polymerase (New England Biolabs), and approximately 50 ng DNA template. LR-PCR amplifications started with an initial denaturation at 94 °C for 2 min followed by 40 cycles of denaturation at 94 °C for 20 s and a combined annealing and elongation step at 68 °C for 9 min and finished with an elongation phase of 15 min at 68 °C. Amplicon quality and quantity were assessed by gel electrophoresis with 10 μL of LR-PCR product analyzed in a 1.0% TAE agarose gel stained with ethidium bromide. After confirmation of successful amplification, remaining reaction volumes were individually purified using a 1.0× ratio of Agencourt AMPure XP beads (Beckman Coulter, Krefeld, Germany) following the manufacturer’s protocol and concentrations were spectrophotometrically quantified using a NanoDrop ND-1000 (Thermo Scientific, Waltham, MA, USA). Equimolar amounts of each amplicon were individually barcoded using PacBio barcoded adapters for multiplex sequencing (Pacific Biosciences, Menlo Park, CA, USA) and used to construct SMRT cell libraries according to the manufacturer’s instructions. Multiplexed libraries consisting of 35 individually barcoded amplicons were subjected to two DNA damage repair steps and exonuclease treatment. After quantification with an Agilent DNA 12000 Kit and the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), the DNA/Polymerase Binding Kit P6 v2 (Pacific Biosciences) and the MagBead Kit v2 (Pacific Biosciences) were used to prepare libraries for sequencing on the PacBio RS II System (Pacific Biosciences). A total of two SMRT cells were sequenced, yielding 98,488 and 101,485 polymerase reads, respectively. Determination of phased consensus sequences was accomplished by the Long Amplicon Analysis (LAA) as implemented in the SMRT Analysis version 2.3.0 followed by manual trimming of primer sequences. As mentioned above, LR-PCR amplifications were performed twice for each of the 35 gorilla samples. Number of subreads per individual totaled from 829 to 2000 with an average of 1639 ± 424. Per individual and replicate, number of subreads ranged from 411 to 1000 (mean 882 ± 185). Subread coverage of consensus sequences was between 118 and 500 (mean 441 ± 106). The estimated accuracy of subreads was high with an average of 99.989% (± 0.0002, range 99.863–99.994%). Comparison of consensus sequences between replicates revealed complete identity. Novel full-length genomic coding sequences were submitted to the MHC-IPD NHP database and officially designated by the nonhuman primate MHC nomenclature committee (Maccari et al. 2017).
Phylogenetic analyses and neutrality tests
Due to their close evolutionary history, we downloaded from the IPD-IMGT/HLA database full-length HLA-B and HLA-C coding region sequences representative of the major allelic lineages (Fukami-Kobayashi et al. 2005; Robinson et al. 2015). Full-length coding region sequences of the chimpanzee (Patr-B and -C), bonobo (Papa-B and -C), gorilla (Gogo-B, -C and Gobe-B, -C), and orangutan (Popy-B, -C, and Poab-B, -C) were retrieved from the IPD-MHC database (Maccari et al. 2017). Using the program MAFFT (Katoh et al. 2002), we generated an alignment which was used to reconstruct a neighbor-joining tree based on the Tamura-Nei model with 500 bootstrap replicates as implemented in MEGA version 6 (Tamura et al. 2013). In addition, we used the program RDP version 4 (Martin et al. 2015) for the detection and identification of recombination events in combination with domain-by-domain phylogenetic analyses to infer evolutionary relationships between Gogo class I genes (Hans et al. 2017). From the contig (000730_F), we extracted the extended gene sequence of the putative Gogo-C gene. Available genomic sequences containing the MHC-B or -C gene of human, chimpanzee, bonobo, gorilla, and orangutan were downloaded from the NCBI GenBank nucleotide database. Using rhesus macaque as outgroup, genomic sequences of Mamu-B4, -B8, and -B9 were obtained from the macaque MHC class I region (GenBank ID: AB128049) (Kulski et al. 2004). Extended gene sequences were aligned using MAFFT and manually edited. Large insertions (≥ 100-bp) identified to be unique to a single sequence or macaque-specific were excluded (Abi-Rached et al. 2011). Following the RDP analysis, the alignment was divided into five segments, each of which was used to generate a phylogeny as described above.
We calculated homozygosity rates at individual gorilla MHC class I genes by counting the number of individuals being homozygous for a given locus divided by the total number of individuals analyzed. However, it has to be noted that gorillas have a comparatively complex MHC class I haplotype structure, in particular the Gogo class I A region exhibits haplotypic variation in gene number and content (Hans et al. 2017). As such, the gorilla MHC class I haplotype containing Gogo-Oko is characterized by the absence of the Gogo-A gene (Gleimer et al. 2011; Hans et al. 2017). Thus, given the potential hemizygous state of Gogo-A and -Oko, homozygosity rates at these loci were calculated using the number of homozygous individuals at Gogo-A or -Oko divided by the total number of individuals possessing this haplotype, respectively.
To detect departures from neutrality in the allele frequency distribution of Gogo class I genes, we used the program DnaSP version 5.10 (Librado and Rozas 2009) to perform the Tajima’s D (Tajima 1989), Fu and Li’s D, and Fu and Li’s F tests (Fu and Li 1993) on each locus separately.
Results and discussion
Previously, we presented a detailed description of the MHC class I genes in gorillas. The combination of LR-PCR amplifications and long-read sequencing technology allowed us to characterize gorilla MHC class I genes in full-length. This approach demonstrated the ability to identify many novel gorilla MHC class I alleles as well as two previously undetected genes, thus substantially contributing to the understanding of MHC class I gene variation in this great ape species (Hans et al. 2017). Nonetheless, crucial for a comprehensive assessment of the MHC diversity is the design of primers reliably amplifying the entire allelic variation at each locus. However, in non-model organisms, such as the gorilla, extensive genomic resources or detailed information about the organization of the MHC are often not available (Babik 2010; but see Wilming et al. 2013). Although an increasing number of whole-genome studies were applied to non-model organisms, including gorillas, the nature of short-read sequencing technologies has prevented an in-depth analysis of the MHC due to its inherent complexity (Scally et al. 2012, 2013; Prado-Martinez et al. 2013; Xue et al. 2015). In contrast, the recent study by Gordon et al. (2016) utilized long-read sequencing technology to generate a high-quality assembly of the gorilla genome which substantially exceeds previously released draft genomes in accuracy and continuity. Analysis of this gorilla assembly suggests that we may have underestimated variation at Gogo-C, the gorilla orthologue of HLA-C, in our previous study (Hans et al. 2017). Thus, we here present the resequencing of Gogo-C aimed towards a more comprehensive description of the gorilla MHC class I gene diversity.
Recombination has shaped the divergent Gogo-C*03:01 allele
Among the 35 gorilla individuals reanalyzed, we obtained 13 genomic sequences of which 10 were completely identical to full-length Gogo-C alleles identified in our previous study (Hans et al. 2017). The remaining three full-length genomic sequences that have not been previously described, Gogo-C*03:01:01:01 (Genbank accession number: MF593179), -C*03:01:01:02 (MF593180), and -C*03:01:01:03 (MF593181) were identical in coding region sequences but differed at 1–2 nucleotide positions in noncoding regions. Phylogenetic analysis of full-length coding region sequences clearly segregates Gogo-C*03:01 apart from MHC-C alleles of humans, chimpanzees, bonobos, gorillas, and orangutans (Fig. 1). This phylogenetically divergent Gogo-C allele appears to be most closely related to Gogo-B*07, the additional B-like gene that is present on some gorilla MHC haplotypes (Hans et al. 2017).
Phylogenetic tree constructed from full-length coding region sequences of MHC-B and -C genes. Relevant bootstrap values (≥ 50%) are shown. The novel allele Gogo-C*03:01 is highlighted in bold. Dashed lines in orange, green, and blue indicate the phylogenetic clusters of the orthologous MHC-B, -C, and -B-like genes, respectively. HLA human, Patr Pan troglodytes, Papa Pan paniscus, Gogo Gorilla gorilla, Gobe Gorilla beringei, Poab Pongo abelii, Popy Pongo pygmaeus
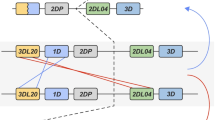
However, intra- and interlocus recombination between members of the MHC class I multigene family can confound orthologous relationships (Jakobsen et al. 1998; Ohta 2010; Hans et al. 2017). Therefore, we investigated extended gene sequences for patterns of recombination in combination with phylogenetic analyses to infer evolutionary relationships between Gogo class I genes (Fig. 2). Domain-by-domain phylogenetic analysis shows that flanking segments of Gogo-C*03:01 are most closely related to MHC-C alleles of humans, chimpanzees, gorillas, and orangutans (Fig. 2a). This relationship extends throughout the gene (Fig. 2b–c) with the exception of the genomic segment encompassing exons 4–8 where Gogo-C*03:01 is most similar to Popy-B*03:02 and Gogo-B*07 alleles (Fig. 2d). In the genomic region downstream of the gene, Gogo-C*03:01 shares sequence similarity with HLA-C, Patr-C, and Gogo-C alleles (Fig. 2e). These findings demonstrate that Gogo-C*03:01 is a divergent Gogo-C allele which through recombination obtained segments from Gogo-B*07, the additional Gogo-B gene (Fig. 2f).
Domain-by-domain phylogenetic analysis of the newly identified Gogo-C*03:01 alleles. a–e Neighbor-joining trees constructed from five genomic segments of MHC-B and -C genes corresponding to the segments given in the schematic at the bottom. Shown in the upper left corner is the size of the genomic segment which was used for each phylogenetic reconstruction. Relevant bootstrap values (≥ 70%) are shown. Black triangles represent compressed MHC-B and -C sequences of human (H), chimpanzee (C), bonobo (B), and gorilla (G) with the total number of sequences given in parentheses. Highlighted in red are sequences of Gogo-C*03:01 alleles. Highlighted in blue are sequences of alleles of Gogo-B*07, the additional B-like gene present on some gorilla MHC haplotypes. Other relevant Gogo alleles are in bold. f Summary of domain-by-domain phylogenetic analyses showing the relationships between MHC-B, MHC-B-like, and MHC-C genes of human (H), chimpanzee (C), bonobo (B), gorilla (G), and orangutan (O). Equivalent segments between or within species have the same color. *, outgroup to both MHC-B and MHC-C, ** outgroup to MHC-B, MHC-B-like and MHC-C. HLA human, Patr Pan troglodytes, Papa Pan paniscus, Gobe Gorilla beringei, Gogo Gorilla gorilla, Popy Pongo pygmaeus, Mamu Macaca mulatta
Functional importance of the Gogo-C*03:01 allele
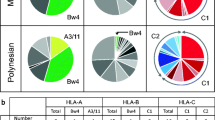
Besides antigen presentation, MHC class I molecules are involved in the regulation of NK cells via KIR. In humans, KIR recognition of HLA class I molecules is determined by amino acid motifs within the α1 domain. Depending on the motif, HLA class I allotypes carry either one of four mutually exclusive epitopes (A3/11, Bw4, C1, and C2) or do not permit KIR recognition (Parham et al. 2012). More specifically, as ligands for lineage II KIR, the Bw4 epitope is carried by HLA-B allotypes with arginine at position 83 while the combination of valine at position 76 and asparagine at position 80 determines the C1 epitope which is recognized by lineage III KIR. In contrast, HLA-C allotypes with asparagine or lysine at position 80 carry the C1 epitope or the C2 epitope, respectively, both of which are ligands for lineage III KIR molecules (reviewed in Parham and Moffett 2013).
It has been shown that certain combinations of MHC class I and KIR variants are associated with resistance and susceptibility to infectious diseases (Kelley and Trowsdale 2005; Parham 2005). These epistatic interactions are considered to have driven the coevolution of the MHC class I genes and their cognate KIR (reviewed in Guethlein et al. 2015). As such, the comparatively low polymorphism and cell surface expression of HLA-C has been attributed to its specialized function as the dominant ligand for KIR (Single et al. 2007; Older Aguilar et al. 2010). Both humans and chimpanzees harbor C1- and C2-bearing MHC-C allotypes whereas gorilla MHC-C allotypes with the C1 epitope have yet to be identified (Adams and Parham 2001; Moesta et al. 2009). Indeed, previously, we found that the C1 epitope is carried only by certain gorilla MHC-B allotypes, in particular Gogo-B*07, whereas all identified gorilla MHC-C allotypes have the C2 epitope. Here, we show that the newly identified gorilla class I allotype, Gogo-C*03:01, exclusively carries the C1 epitope at Gogo-C, as summarized in Fig. 3. We identified this allotype in 17 of the 35 gorillas analyzed, suggesting its important function in controlling NK cell responses via KIR (Table 1).
Motifs encoding KIR ligands in the sequences of Gogo-C molecules. Identity to consensus sequence is denoted by a dash. For differences from the consensus sequence, the amino acid is shown. The newly identified Gogo-C*03:01 allotype is highlighted in bold. Positions highlighted in blue and red define the C1 and the C2 epitope, respectively
Although we cannot completely rule out the possibility that we might have missed additional alleles, several lines of evidence indicate that we have identified the complete allelic diversity at Gogo class I genes: (i) homozygosity levels between Gogo class I genes are not substantially different with 18.7, 18.7, 17.1, and 14.2% of individuals homozygous at Gogo-A, -Oko, -B, and -C, respectively; (ii) many homozygous individuals have only one allele at two or more loci, presumably due to strong linkage disequilibrium between Gogo class I genes; and (iii) alleles identified in related individuals do not show deviations from expected inheritance patterns (Table 1). Thus, we are confident that we herewith fully complement the characterization of allelic diversity at Gogo class I genes.
A selective sweep in gorillas?
Recent genome-wide surveys of gorilla genetic variation reported a marked reduction of diversity close to the MHC suggestive of a recent selective sweep (Scally et al. 2013; Xue et al. 2015). Indeed, we have previously shown that the overall Gogo class I gene diversity is low relative to humans and chimpanzees despite gorillas having a comparatively complex MHC class I haplotype structure (Hans et al. 2017). Further evidence suggesting that gorillas experienced a reduction of their MHC class I repertoire comes from the uneven allele frequency distribution of Gogo class I genes, as illustrated in Fig. 4. Among the 33 unrelated gorillas analyzed, we show that the frequencies of the two most common Gogo-A alleles, Gogo-A*01:01 and -A*04:01, account for over 60% of the total variance (Fig. 4a). For the Gogo-Oko haplotype, the allele Gogo-Oko*01:01 has the highest proportion of the total allele frequency (15%) (Fig. 4a). The absence of a predominant allele lineage at Gogo-B, the gorilla orthologue of HLA-B, may be a result of frequent recombination promoting the exchange of sequence motifs at this locus (McAdam et al. 1994; Parham and Ohta 1996; Jakobsen et al. 1998). Indeed, phylogenetic analyses revealed the prevalence of divergent gorilla B locus alleles with many of them present at frequencies below or equal to 5%, suggesting the rapid diversification of Gogo-B alleles (Fig. 4b) (Hans et al. 2017). In contrast, the frequency spectrum at Gogo-C is also skewed towards high-frequency alleles: the Gogo-C*01:01 allele has a frequency of 39% whereas the novel Gogo-C*03:01 allele was identified in 16 of the 33 unrelated gorillas and, hence, accounts for 24% of the total allele frequency (Fig. 4c). However, unlike a selective sweep in which, by definition, all adaptive alleles arise from a single mutation and coalesce into a single ancestral lineage, gorillas have maintained several allelic lineages at individual Gogo class I genes (Wilson et al. 2014; Hans et al. 2017). In accordance, the neutrality tests applied do not indicate an excess of rare alleles characteristic of a selective sweep (Tajima 1989; Kim 2006). More specifically, with the exception of Gogo-C, Tajima’s D shows no significant deviations from neutral expectations whereas Fu and Li’s D and F statistics indicate a skew in the allele frequency distribution among Gogo class I genes (Table 2). Significantly positive values reflect a deficit of rare alleles which can result from balancing selection, population structure, and/or bottlenecks (Jensen et al. 2005; Biswas and Akey 2006).
Balancing selection maintains advantageous diversity in the population and is considered to be one of the main mechanisms maintaining MHC polymorphism over long evolutionary time periods, a process known as trans-species polymorphism (Bernatchez and Landry 2003; Klein et al. 2007). Polymorphism of MHC class I genes is predominantly confined to exons 2 and 3, encoding the contact residues of the antigen-binding site (ABS), typically showing elevated ratios of nonsynonymous (Ka) to synonymous (Ks) substitution rates indicative of balancing selection (Hughes and Nei 1988; Ohta 1991; Hughes and Yeager 1998; Spurgin and Richardson 2010). Previously, we analyzed Ka and Ks substitution rates within ABS of Gogo class I genes (Hans et al. 2017). Although significant differences between Ka and Ks were observed only for Gogo-B, patterns of Ka/Ks ratios suggest that balancing selection acts to maintain diversity within ABS of gorilla MHC class I genes. However, except for Gogo-B, absolute numbers of variable sites within MHC class I alleles are substantially lower in gorillas than in chimpanzees and humans. Indeed, as reflected by an interspecies comparison, there is only limited allelic variation within lineages of gorilla MHC class I genes including Gogo-B (Hans et al. 2017). Similarly, differences between newly identified Gogo-C*03:01 alleles are only due to single nucleotide substitutions in noncoding regions. Furthermore, many allelic lineages that are present in chimpanzees and/or humans are absent in gorillas, contrary to expectations given the trans-species mode of evolution (Trowsdale 1995; Klein et al. 2007; Hans et al. 2017). Thus, although balancing selection may be acting on short evolutionary time scales, the overall patterns observed at Gogo class I genes are also suggestive of ancient and/or recent demographic changes in gorillas.
Although previous studies revealed substructure within gorilla populations, particularly within western lowland gorillas, these findings are unlikely to explain the high homogeneity of Gogo class I alleles identified in our samples (Clifford et al. 2004; Thalmann et al. 2007; Anthony et al. 2007; Scally et al. 2013; Fünfstück et al. 2014). However, it has to be noted that we cannot completely exclude the possibility of sampling from genetically homogeneous populations, despite the knowledge of geographical origins of most gorillas (Hans et al. 2017). Nevertheless, it has been suggested that the present-day substructure observed within western lowland gorillas rather reflects ancestral population structure (Thalmann et al. 2007). Postglacial forest recovery during the Pleistocene allowed the expansion and admixture between previously separated populations, thereby promoting genetic diversity (Anthony et al. 2007; Thalmann et al. 2007; Scally et al. 2013).
Indeed, although western lowland gorillas show the highest levels of genetic diversity relative to other great ape species, there are indications of a severe genetic bottleneck (Prado-Martinez et al. 2013). Recent demographic inferences revealed that western lowland gorillas experienced a fourfold decrease in their effective population size (N e ) approximately 30,000–50,000 years ago (Prado-Martinez et al. 2013; McManus et al. 2015; Gordon et al. 2016). In accordance, distributions of genomic runs of homozygosity (ROH) in gorillas are indicative of a history of inbreeding with patterns similar to those in human populations that have undergone strong genetic bottlenecks (Kirin et al. 2010; Prado-Martinez et al. 2013; Xue et al. 2015). Furthermore, consistent with our observations at Gogo class I genes, genome-wide analysis of the allele frequency spectrum revealed a deficit of rare alleles, further indicating that western lowland gorillas experienced a recent episode of population contraction (Scally et al. 2013). In accordance, alleles at Gogo class I genes accumulated only limited polymorphisms within both coding and noncoding regions (Hans et al. 2017). Indeed, previous estimates inferred that the western lowland gorilla population has decreased by more than 60% in the past 20–25 years (Vogel 2007). Besides habitat fragmentation and degradation, major threats responsible for the drastic decline of gorilla populations include poaching and infectious diseases, in particular Ebola (Walsh et al. 2003; Junker et al. 2012). In some regions, the Ebola epidemic has killed more than 90% of the western lowland gorillas, which has prompted their conservation status change to “critically endangered” (Bermejo et al. 2006; Hopkin 2007; Le Gouar et al. 2009). Even more dramatic, recent census estimates predict a decline of western gorillas by over 80% in the next three generations, a scenario which would bring gorillas to the brink of extinction (Maisels et al. 2016).
Taken together, these findings strongly suggest that the low variation observed at Gogo class I genes resulted from successive genetic bottlenecks that caused the loss of MHC diversity in gorillas. Following this scenario, we propose that after the initial reduction of the Gogo class I repertoire, functionally divergent alleles were maintained in the population through balancing selection, as can be seen from the comparatively high nucleotide diversity values (Hans et al. 2017). Indeed, similar patterns have been observed in other species that have undergone severe bottlenecks (Hedrick et al. 2000a, 2000b; Sommer 2005; Ellison et al. 2012). Subsequently, however, Gogo class I diversity became further decreased during the recent bottleneck(s), as reflected by the low polymorphisms that accumulated within allelic lineages of Gogo class I genes.
In sum, we demonstrated the advantage of high-quality genomes for a comprehensive description of the MHC diversity in non-model organisms, such as the gorilla. Reassessment of the Gogo-C diversity showed that gorillas, unlike previous suggestions, exhibit a high-frequency allele exclusively encoding a Gogo-C allotype with the C1 epitope. Furthermore, we propose that the low diversity at Gogo class I genes can be best explained by drastic demographic changes gorillas experienced in the ancient and recent past. However, we strongly encourage future studies to incorporate more sensitive haplotype-based analyses to test this hypothesis.
References
Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, Kimani J, Carrington M, Middleton D, Rajalingam R, Beksac M, Marsh SG, Maiers M, Guethlein LA, Tavoularis S, Little AM, Green RE, Norman PJ, Parham P (2011) The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334:89–94. https://doi.org/10.1126/science.1209202
Adams EJ, Parham P (2001) Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev 183:41–64
Anthony NM, Johnson-Bawe M, Jeffery K, Clifford SL, Abernethy KA, Tutin CE, Lahm SA, White LJ, Utley JF, Wickings EJ, Bruford MW (2007) The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc Natl Acad Sci U S A 104:20432–20436. https://doi.org/10.1073/pnas.0704816105
Babik W (2010) Methods for MHC genotyping in non-model vertebrates. Mol Ecol Resour 10:237–251. https://doi.org/10.1111/j.1755-0998.2009.02788.x
Bermejo M, Rodríguez-Teijeiro JD, Illera G, Barroso A, Vilà C, Walsh PD (2006) Ebola outbreak killed 5000 gorillas. Science 314:1564. https://doi.org/10.1126/science.1133105
Biswas S, Akey JM (2006) Genomic insights into positive selection. Trends Genet 22:437–446. https://doi.org/10.1016/j.tig.2006.06.005
Bernatchez L, Landry C (2003) MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J Evol Biol 16:363–377
Bontrop RE (2006) Comparative genetics of MHC polymorphisms in different primate species: duplications and deletions. Hum Immunol 67:388–397. https://doi.org/10.1016/j.humimm.2006.03.007
Clifford SL, Anthony NM, Bawe-Johnson M, Abernethy KA, Tutin CE, White LJ, Bermejo M, Goldsmith ML, McFarland K, Jeffery KJ, Bruford MW, Wickings EJ (2004) Mitochondrial DNA phylogeography of western lowland gorillas (Gorilla gorilla gorilla). Mol Ecol 13(1551–1565):1567. https://doi.org/10.1111/j.1365-294X.2004.02140.x
Davis MM, Bjorkman PJ (1988) T-cell antigen receptor genes and T-cell recognition. Nature 334:395–402. https://doi.org/10.1038/334395a
Ellison A, Allainguillaume J, Girdwood S, Pachebat J, Peat KM, Wright P, Consuegra S (2012) Maintaining functional major histocompatibility complex diversity under inbreeding: the case of a selfing vertebrate. Proc Biol Sci 279:5004–5013. https://doi.org/10.1098/rspb.2012.1929
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fünfstück T, Arandjelovic M, Morgan DB, Sanz C, Breuer T, Stokes EJ, Reed P, Olson SH, Cameron K, Ondzie A, Peeters M, Kühl HS, Cipolletta C, Todd A, Masi S, Doran-Sheehy DM, Bradley BJ, Vigilant L (2014) The genetic population structure of wild western lowland gorillas (Gorilla gorilla gorilla) living in continuous rain forest. Am J Primatol 76:868–878. https://doi.org/10.1002/ajp.22274
Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, Tateno Y (2005) Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A 102:9230–9234. https://doi.org/10.1073/pnas.0500770102
Gleimer M, Wahl AR, Hickman HD, Abi-Rached L, Norman PJ, Guethlein LA, Hammond JA, Draghi M, Adams EJ, Juo S, Jalili R, Gharizadeh B, Ronaghi M, Garcia KC, Hildebrand WH, Parham P (2011) Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A*02 have overlapping peptide-binding repertoires. J Immunol 186:1575–1588. https://doi.org/10.4049/jimmunol.1002990
Gordon D, Huddleston J, Chaisson MJ, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, Dunn C, Baker C, Armstrong J, Diekhans M, Paten B, Shendure J, Wilson RK, Haussler D, Chin CS, Eichler EE (2016) Long-read sequence assembly of the gorilla genome. Science 352:aae0344. https://doi.org/10.1126/science.aae0344
Guethlein LA, Norman PJ, Hilton HG, Parham P (2015) Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev 267:259–282. https://doi.org/10.1111/imr.12326
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hans JB, Bergl RA, Vigilant L (2017) Gorilla MHC class I gene and sequence variation in a comparative context. Immunogenetics 69:303–323. https://doi.org/10.1007/s00251-017-0974-x
Hedrick PW, Lee RN, Parker KM (2000a) Major histocompatibility complex (MHC) variation in the endangered Mexican wolf and related canids. Heredity (Edinb) 85:617–624
Hedrick PW, Parker KM, Gutiérrez-Espeleta GA, Rattink A, Lievers K (2000b) Major histocompatibility complex variation in the Arabian oryx. Evolution 54:2145–2151
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170. https://doi.org/10.1038/335167a0
Hughes AL, Nei M (1989) Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol Biol Evol 6:559–579
Hughes AL, Yeager M (1998) Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet 32:415–435. https://doi.org/10.1146/annurev.genet.32.1.415
Hopkin M (2007) Gorillas on the list. Nature 449:127. https://doi.org/10.1038/449127a
Jakobsen IB, Wilson SR, Easteal S (1998) Patterns of reticulate evolution for the classical class I and II HLA loci. Immunogenetics 48:312–323
Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD (2005) Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170:1401–1410. https://doi.org/10.1534/genetics.104.038224
Junker J, Blake S, Boesch C, Campbell G, Toit LD, Duvall C, Ekobo A, Etoga G, Galat-Luong A, Gamys J, Ganas-Swaray J, Gatti S, Ghiurghi A, Granier N, Hart J, Head J, Herbinger I, Hicks TC, Huijbregts B, Imong IS, Kuempel N, Lahm S, Lindsell J, Maisels F, McLennan M, Martinez L, Morgan B, Morgan D, Mulindahabi F, Mundry R, N’Goran KP, Normand E, Ntongho A, Okon DT, Petre CA, Plumptre A, Rainey H, Regnaut S, Sanz C, Stokes E, Tondossama A, Tranquilli S, Sunderland-Groves J, Walsh P, Warren Y, Williamson EA, Kuehl HS (2012) Recent decline in suitable environmental conditions for African great apes. Diversity Distrib 18:1077–1091. https://doi.org/10.1111/ddi.12005
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kelley J, Trowsdale J (2005) Features of MHC and NK gene clusters. Transpl Immunol 14:129–134. https://doi.org/10.1016/j.trim.2005.03.001
Kelley J, Walter L, Trowsdale J (2005) Comparative genomics of major histocompatibility complexes. Immunogenetics 56:683–695. https://doi.org/10.1007/s00251-004-0717-7
Kim Y (2006) Allele frequency distribution under recurrent selective sweeps. Genetics 172:1967–1978. https://doi.org/10.1534/genetics.105.048447
Kirin M, McQuillan R, Franklin CS, Campbell H, McKeigue PM, Wilson JF (2010) Genomic runs of homozygosity record population history and consanguinity. PLoS One 5:e13996. https://doi.org/10.1371/journal.pone.0013996
Klein J, Sato A, Nikolaidis N (2007) MHC, TSP, and the origin of species: from immunogenetics to evolutionary genetics. Annu Rev Genet 41:281–304. https://doi.org/10.1146/annurev.genet.41.110306.130137
Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H (2002) Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev 190:95–122
Kulski JK, Anzai T, Shiina T, Inoko H (2004) Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol 21:2079–2091. https://doi.org/10.1093/molbev/msh216
Laehnemann D, Borkhardt A, McHardy AC (2016) Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinform 17:154–179. https://doi.org/10.1093/bib/bbv029
Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23:225–274. https://doi.org/10.1146/annurev.immunol.23.021704.115526
Lawlor DA, Warren E, Taylor P, Parham P (1991) Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I. J Exp Med 174:1491–1509
Le Gouar PJ, Vallet D, David L, Bermejo M, Gatti S, Levréro F, Petit EJ, Ménard N (2009) How Ebola impacts genetics of Western lowland gorilla populations. PLoS One 4:e8375. https://doi.org/10.1371/journal.pone.0008375
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, Ho CS, de Groot NG, Flicek P, Bontrop RE, Hammond JA, Marsh SGE (2017) IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45:D860–D864. https://doi.org/10.1093/nar/gkw1050
Maisels F, Bergl RA, Williamson EA (2016) Gorilla gorilla (errata version published in 2016). The IUCN Red List of Threatened Species 2016. https://doi.org/10.2305/IUCN.UK.2016-2.RLTS.T9404A17963949.en. Assessed 28 June 2017
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: https://doi.org/10.1093/ve/vev003
Martínez-Laso J, Gómez-Casado E, Arnaiz-Villena A (2006) Description of seven new non-human primate MHC-B alleles. Tissue Antigens 67:85–88. https://doi.org/10.1111/j.1399-0039.2005.00496.x
McAdam SN, Boyson JE, Liu X, Garber TL, Hughes AL, Bontrop RE, Watkins DI (1994) A uniquely high level of recombination at the HLA-B locus. Proc Natl Acad Sci U S A 91:5893–5897
McManus KF, Kelley JL, Song S, Veeramah KR, Woerner AE, Stevison LS, Ryder OA, Ape Genome Project G, Kidd JM, Wall JD, Bustamante CD, Hammer MF (2015) Inference of gorilla demographic and selective history from whole-genome sequence data. Mol Biol Evol 32:600–612. https://doi.org/10.1093/molbev/msu394
Moesta AK, Abi-Rached L, Norman PJ, Parham P (2009) Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol 182:3628–3637. https://doi.org/10.4049/jimmunol.0803401
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. https://doi.org/10.1146/annurev.genet.39.073003.112240
Ohta T (1991) Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc Natl Acad Sci U S A 88:6716–6720
Ohta T (2010) Gene conversion and evolution of gene families: an overview. Genes 1:349–356. https://doi.org/10.3390/genes1030349
Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P (2010) Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol 185:4238–4251. https://doi.org/10.4049/jimmunol.1001494
Parham P (2005) MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 5:201–214. https://doi.org/10.1038/nri1570
Parham P, Moffett A (2013) Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol 13:133–144. https://doi.org/10.1038/nri3370
Parham P, Ohta T (1996) Population biology of antigen presentation by MHC class I molecules. Science 272:67–74
Parham P, Norman PJ, Abi-Rached L, Guethlein LA (2012) Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond Ser B Biol Sci 367:800–811. https://doi.org/10.1098/rstb.2011.0266
Piontkivska H, Nei M (2003) Birth-and-death evolution in primate MHC class I genes: divergence time estimates. Mol Biol Evol 20:601–609. https://doi.org/10.1093/molbev/msg064
Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prüfer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubí C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, Siegismund H, Blanc J, Agueda-Calpena L, Gut M, Fulton L, Tishkoff SA, Mullikin JC, Wilson RK, Gut IG, Gonder MK, Ryder OA, Hahn BH, Navarro A, Akey JM, Bertranpetit J, Reich D, Mailund T, Schierup MH, Hvilsom C, Andrés AM, Wall JD, Bustamante CD, Hammer MF, Eichler EE, Marques-Bonet T (2013) Great ape genetic diversity and population history. Nature 499:471–475. https://doi.org/10.1038/nature12228
Robinson J, Halliwell JA, Hayhurst JH, Flicek P, Parham P, Marsh SGE (2015) The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 43:D423–D431. https://doi.org/10.1093/nar/gku1161
Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB (2013) Characterizing and measuring bias in sequence data. Genome Biol 14:R51. https://doi.org/10.1186/gb-2013-14-5-r51
Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, Hobolth A, Lappalainen T, Mailund T, Marques-Bonet T, McCarthy S, Montgomery SH, Schwalie PC, Tang YA, Ward MC, Xue Y, Yngvadottir B, Alkan C, Andersen LN, Ayub Q, Ball EV, Beal K, Bradley BJ, Chen Y, Clee CM, Fitzgerald S, Graves TA, Gu Y, Heath P, Heger A, Karakoc E, Kolb-Kokocinski A, Laird GK, Lunter G, Meader S, Mort M, Mullikin JC, Munch K, O'Connor TD, Phillips AD, Prado-Martinez J, Rogers AS, Sajjadian S, Schmidt D, Shaw K, Simpson JT, Stenson PD, Turner DJ, Vigilant L, Vilella AJ, Whitener W, Zhu B, Cooper DN, de Jong P, Dermitzakis ET, Eichler EE, Flicek P, Goldman N, Mundy NI, Ning Z, Odom DT, Ponting CP, Quail MA, Ryder OA, Searle SM, Warren WC, Wilson RK, Schierup MH, Rogers J, Tyler-Smith C, Durbin R (2012) Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. https://doi.org/10.1038/nature10842
Scally A, Yngvadottir B, Xue Y, Ayub Q, Durbin R, Tyler-Smith C (2013) A genome-wide survey of genetic variation in gorillas using reduced representation sequencing. PLoS One 8:e65066. https://doi.org/10.1371/journal.pone.0065066
Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M (2007) Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 39:1114–1119. https://doi.org/10.1038/ng2077
Sommer S (2005) The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool 2:16. https://doi.org/10.1186/1742-9994-2-16
Spurgin LG, Richardson DS (2010) How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci 277:979–988. https://doi.org/10.1098/rspb.2009.2084
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Thalmann O, Fischer A, Lankester F, Pääbo S, Vigilant L (2007) The complex evolutionary history of gorillas: insights from genomic data. Mol Biol Evol 24:146–158. https://doi.org/10.1093/molbev/msl160
Trowsdale J (1995) “Both man & bird & beast”: comparative organization of MHC genes. Immunogenetics 41:1–17
Urvater JA, Hickman H, Dzuris JL, Prilliman K, Allen TM, Schwartz KJ, Lorentzen D, Shufflebotham C, Collins EJ, Neiffer DL, Raphael B, Hildebrand W, Sette A, Watkins DI (2001) Gorillas with spondyloarthropathies express an MHC class I molecule with only limited sequence similarity to HLA-B27 that binds peptides with arginine at P2. J Immunol 166:3334–3344
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251. https://doi.org/10.1146/annurev.immunol.20.092501.134942
Vogel G (2007) Conservation. Scientists say Ebola has pushed western gorillas to the brink. Science 317:1484. https://doi.org/10.1126/science.317.5844.1484
Walsh PD, Abernethy KA, Bermejo M, Beyers R, De Wachter P, Akou ME, Huijbregts B, Mambounga DI, Toham AK, Kilbourn AM, Lahm SA, Latour S, Maisels F, Mbina C, Mihindou Y, Obiang SN, Effa EN, Starkey MP, Telfer P, Thibault M, Tutin CE, White LJ, Wilkie DS (2003) Catastrophic ape decline in western equatorial Africa. Nature 422:611–614. https://doi.org/10.1038/nature01566
Watkins DI, Chen ZW, Garber TL, Hughes AL, Letvin NL (1991) Segmental exchange between MHC class I genes in a higher primate: recombination in the gorilla between the ancestor of a human non-functional gene and an A locus gene. Immunogenetics 34:185–191
Wilming LG, Hart EA, Coggill PC, Horton R, Gilbert JG, Clee C, Jones M, Lloyd C, Palmer S, Sims S, Whitehead S, Wiley D, Beck S, Harrow JL (2013) Sequencing and comparative analysis of the gorilla MHC genomic sequence. Database (Oxford) 2013:bat011. doi: https://doi.org/10.1093/database/bat011
Wilson BA, Petrov DA, Messer PW (2014) Soft selective sweeps in complex demographic scenarios. Genetics 198:669–684. https://doi.org/10.1534/genetics.114.165571
Wong P, Pamer EG (2003) CD8 T cell responses to infectious pathogens. Annu Rev Immunol 21:29–70. https://doi.org/10.1146/annurev.immunol.21.120601.141114
Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN, de Manuel M, Hernandez-Rodriguez J, Lobon I, Siegismund HR, Pagani L, Quail MA, Hvilsom C, Mudakikwa A, Eichler EE, Cranfield MR, Marques-Bonet T, Tyler-Smith C, Scally A (2015) Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348:242–245. https://doi.org/10.1126/science.aaa3952
Acknowledgements
This work as well as open access was funded by the Max Planck Society.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 133 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hans, J.B., Vigilant, L. Discovery of gorilla MHC-C expressing C1 ligand for KIR. Immunogenetics 70, 293–304 (2018). https://doi.org/10.1007/s00251-017-1038-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-017-1038-y