Abstract
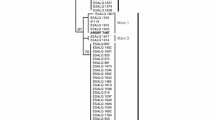
We studied the species diversity and population genetic structure of isolates of fungi from the entomopathogenic genus Metarhizium that had been isolated from sugarcane crops and surrounding grass. Soil and leaf samples were taken on four sampling occasions over 13 months (October 2014–October 2015). Isolations were made using the Galleria mellonella baiting method and selective media. Phylogenetic placement of isolates was done by sequencing a fragment of the 5′ of the elongation factor 1-α gene (EF1-α). Population genetic structure was determined by analysing this sequence information using AMOVA and Haplotype network analyses. Genotypic diversity was studied using microsatellite genotyping. The most abundant species was M. anisopliae s.s. (80 isolates), then M. pingshaense (three isolates), and M. guizhouense (one isolate). More than 50% of the genetic variation was explained by the time the samples were collected regardless of plant host association. Some haplotypes were found on the first sampling date and then not found on subsequent sampling dates, while other haplotypes were found initially, disappeared, but then found again on the last sampling date. To the best of our knowledge, this is the first report of the population genetic structure of M. anisopliae species in time and space. The effect of abiotic factors is discussed.





Similar content being viewed by others
References
Meyling NV, Eilenberg J (2007) Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agroecosystems: potential for conservation biological control. Biol. Control 43:145–155
de Faria MR, Wraight SP (2007) Mycoinsecticides and Mycoacaricides: a comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 43:237–256
Vega FE, Goettel MS, Blackwell M, Chandler D, Jackson MA, Keller S, Koike M, Maniania NK, Monzon A, Ownley BH, Pell JK (2009) Fungal entomopathogens: new insights on their ecology. Fungal Ecol. 2:149–159
Roy HE, Brodie EL, Chandler D, Goettel MS, Pell JK, Wajnberg E, Vega FE (2010) Deep space and hidden depths: understanding the evolution and ecology of fungal entomopathogens. BioControl 55:1–6
Meyling NV, Lübeck M, Buckley EP, Eilenberg J, Rehner SA (2009) Community composition, host range and genetic structure of the fungal entomopathogen Beauveria in adjoining agricultural and seminatural habitats. Mol. Ecol. 18:1282–1293
Meyling NV, Pilz C, Keller S, Widmer F, Enkerli J (2012) Diversity of Beauveria spp. isolates from pollen beetles Meligethes aeneus in Switzerland. J Invertbr Pathol 109:76–82
Kepler RM, Ugine TA, Maul JE, Cavigelli MA, Rehner SA (2015) Community composition and population genetics of insect pathogenic fungi in the genus Metarhizium from soils of a long-term agricultural research system. Environ. Microbiol. 17:2791–2804
Ormond EL, Thomas APM, Pugh PJA, Pell JK, Roy HE (2010) A fungal pathogen in time and space: the population dynamics of Beauveria bassiana in a conifer forest. FEMS Microb Ecol 74:146–154
Rezende JM, Riguetti Zanardo AB, Lopes MS, Delalibera Jr I, Rehner SA (2015) Phylogenetic diversity of Brazilian Metarhizium associated with sugarcane agriculture. BioControl 60:495–505
Bidochka MJ, Small CL (2005) Phylogeography of Metarhizium, an insect pathogenic fungus. In: Vega FE, Blackwell M (eds) Insect-fungal associations ecology and evolution. University Press, Oxford, pp. 28–50
Bischoff JF, Rehner SA, Humber RA (2009) A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 101:512–530
Vega FE, Meyling NV, Luangsa-ard JJ, Blackwell M (2012) Fungal entomopathogens. In: Vega FE, Kaya HK (eds) Insect pathology, 2nd edn. Academic Press, London, pp. 171–219
Hernández-Domínguez C, Guzmán-Franco AW, Carrillo-Benítez MG, Alatorre-Rosaa R, Rodriguez-Leyva E, Villanueva-Jiménez JA (2016a) Specific diversity of Metarhizium isolates infecting Aeneolamia spp. (Hemiptera: Cercopidae) in sugarcane plantations. Neotrop Entomol 45:80–87
Wyrebek M, Huber C, Sasan RK, Bidochka MJ (2011) Three sympatrically occurring species of Metarhizium show plant rhizosphere specificity. Microbiology 157:2904–2911
Zimmermann G (1986) The Galleria bait method for detection of entomopathogenic fungi in soil. J. Appl. Entomol. 102:213–215
Dolci P, Guglielmo F, Secchi F, Ozino OI (2006) Persistence and efficacy of Beauveria brongniartii strains applied as biocontrol agents against Melolontha melolontha in the valley of Aosta (Northwest Italy). J. Appl. Microbiol. 100:1063–1072
Keller S, Kessler P, Schweizer C (2003) Distribution of insect pathogenic soil fungi in Switzerland with special reference to Beauveria brongniartii and Metharhizium anisopliae. BioControl 48:307–319
Vänninen I, Tyni-Juslin J, Hokkanen H (2000) Persistence of augmented Metarhizium anisopliae and Beauveria bassiana in Finnish agricultural soils. BioControl 45:201–222
Hernández-Domínguez C, Cerroblanco-Baxcajay ML, Alvarado-Aragón LU, Hernández-López G, Guzmán-Franco AW (2016b) Comparison of the relative efficacy of an insect baiting method and selective media for diversity studies of Metarhizium species in the soil. Biocontrol Sci. Tech. 26:707–717
Fernandes KKE, Keyser AC, Rangel NED, Foster NR, Roberts WD (2010) CTC medium: a novel dodine-free selective medium for isolating entomopathogenic fungi especially Metarhizium acridum from soil. Biol. Control 54:197–205
Rangel NED, Dettenmaier SJ, Fernandes EKK, Roberts DW (2010) Susceptibility of Metarhizium spp. and other entomopathogenic fungi to dodine-based selective media. Biocontrol Sci. Tech. 20:375–389
Humber RA (2012) Identification of entomopathogenic fungi. In: Lacey LA (ed) Manual of techniques in invertebrate pathology, 2nd edn. Academic Press, London, pp. 151–187
Meyling NV, Eilenberg J (2006) Occurrence and distribution of soil borne entomopathogenic fungi within a single organic agroecosystem. Agric. Ecosyst. Environ. 113:336–341
Pérez-González VH, Guzmán-Franco AW, Alatorre-Rosas R, Hernández-López J, Hernández-López A, Carrillo-Benitez MG, Baverstock J (2014) Specific diversity of the entomopathogenic fungi Beauveria and Metarhizium in Mexican agricultural soils. J. Invertebr. Pathol. 119:54–61
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Tamura K, Paterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657–1660
Excoffier L, Lischer HEL (2015) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Oulevey C, Widmer F, Kölliker R, Enkerli J (2009) An optimized microsatellite marker set for detection of Metarhizium anisopliae genotype diversity on field and regional scales. Mycol. Res. 13:1016–1024
Enkerli J, Kölliker R, Keller S, Widmer F (2005) Isolation and characterization of microsatellite markers from the entomopathogenic fungus Metarhizium anisopliae. Mol. Ecol. Notes 5:384–386
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Storza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Park SDE (2001) Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis University of Dublin, Ireland
Nei M, Saito N (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci 12:357–358
Steinwender BM, Enkerli J, Widmer F, Eilenberg J, Kristensen HL, Bodochke MJ, Meyling NV (2015) Root isolations of Metarhizium spp. from crops reflect diversity in the soil and indicate no plant specificity. J. Invertebr. Pathol. 132:142–148
Muñiz-Reyes E, Guzmán-Franco AW, Sánchez-Escudero J, Nieto-Angel R (2014) Occurrence of entomopathogenic fungi in tejocote (Crataegus mexicana) orchard soils and their pathogenicity against Rhagoletis pomonella. J. Appl. Microbiol. 117:1450–1462
Bidochka MJ, Kamp AM, Levander TM, Dekoning J, De Croos JA (2001) Habitat association in two genetic groups of the insect-pathogenic fungus Metarhizium anisopliae: uncovering cryptic species? Appl. Environ. Microbiol. 67:1335–1342
Bruck DJ (2005) Ecology of Metarhizium anisopliae in soilless potting media and the rhizosphere: implications for pest management. Biol. Control 32:155–163
Smith DM, Inman-Bamber NG, Thorburn PJ (2005) Growth and function of the sugarcane root system. Field Crops Res 92:169–183
Laclau PB, Laclau JP (2009) Growth of the whole root system for a plant crop of sugarcane under rainfed and irrigated environments in Brazil. Field Crops 114:351–360
Hu G, St. Leger RJ (2002) Field studies using a recombinant mycoinsecticide (Metarhizium anisopliae) reveal that it is rhizosphere competent. Appl. Environ. Microbiol. 68:6383–6387
Meyling NV, Hajek AE (2010) Principles from community and metapopulation ecology: application to fungal entomopathogens. BioControl 55:39–54
Levin SA (1992) The problem of pattern and scale in ecology. Ecology 73:1943–1967
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57:233–266
Brady NC, Weil RR (1999) The nature and property of soils. Prentice Hall, Upper Saddle Hall, NJ
Schneider S, Widmer F, Jacot K, Kölliker R, Enkerli J (2012) Spatial distribution of Metarhizium clade 1 in agricultural landscapes with arable land and different semi-natural habitats. Appl. Soil Ecol. 52:20–28
FAO (2015) Crop Water Information: Sugarcane 2015. Available at: http://www.fao.org/nr/water/cropinfo_sugarcane.html [Accessed September 28, 2016]
INIFAP (2016) Red Nacional de Estaciones Agrometeorológicas Automatizadas INIFAP. 2016. Available at: http://clima.inifap.gob.mx/LNMySR/Estaciones/ConsultaDiarios15Min?Estado=29&Estacion=38219 [Accessed September 16, 2016]
Acknowledgements
CHD received and PhD scholarship from CONACyT-Mexico. The authors are grateful to Dr. Stephen Rehner for kindly providing initial training to the first author in the amplification and sequencing of the EF1-α fragment in his laboratory. We are also grateful to Fabián Vázquez Moreno, Jorge Hernandez López and Stephanie Guzman Valencia for their valuable help during sampling, isolate collection and technical assistance for the microsatellite genotyping analysis. The authors are grateful to Drs. R. Alatorre-Rosas, E. Rodriguez-Leyva, M.G. Carrillo-Benitez and J.A. Villanueva-Jimenez for their valuable comments and suggestions during the initial conception of this research. No conflict of interest is declared by the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hernández-Domínguez, C., Guzmán-Franco, A.W. Species Diversity and Population Dynamics of Entomopathogenic Fungal Species in the Genus Metarhizium—a Spatiotemporal Study. Microb Ecol 74, 194–206 (2017). https://doi.org/10.1007/s00248-017-0942-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-0942-x