Abstract
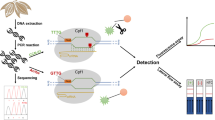
Panax ginseng and Panax quinquefolius, which are commonly called Chinese ginseng and American ginseng respectively, have different medicinal properties and market values; however, these samples can be difficult to differentiate from one another based on physical appearances of the samples especially when they are in powdery or granular forms. A molecular technique is thus needed to overcome this difficulty; this technique is based on the nucleic acid test (NAT) conducted on the microfluidic chip surface. Three single nucleotide polymorphism (SNP) sites (i.e. N1, N2, N3) on the Panax genome that differ between P. ginseng (G) and P. quinquefolius (Q) have been selected to design probes for the NAT. Primers were designed to amplify the antisense strands by asymmetric PCR. We have developed three different NAT methodologies involving surface immobilization and subsequent (stop flow or dynamic) hybridization of probes (i.e. N1G, N1Q, N2G, N2Q, N3Q) to the antisense strands. These NAT methods consist of two steps, namely immobilization and hybridization, and each method is distinguished by what is immobilized on the microfluidic chip surface in the first step (i.e. probe, target or capture strand). These three NATs developed are called probe-target method 1, target-probe method 2 and three-strand complex method 3. Out of the three methods, it was found that the capture strand-target-probe method 3 provided the best differentiation of the ginseng species, in which a 3' NH2 capture strand is first immobilized and the antisense PCR strand is then bound, while N2G and N3Q probes are used for detection of P. ginseng (G) and P. quinquefolius (Q) respectively.
Graphical Abstract









Similar content being viewed by others
References
Hu W, Liu N, Tian Y, Zhang L. Molecular cloning, expression, purification, and functional characterization of dammarenediol synthase from Panax ginseng. Biomed Res Int. 2012;2013:1–7.
Shibata S, Tanaka O, Sôma K, Iida Y, Ando T, Nakamura H. Studies on saponins and sapogenins of ginseng the structure of panaxatriol. Tetrahedron Lett. 1965;6:207–13.
Fulder SJ. The growth of cultured human fibroblasts treated with hydrocortisone and extracts of the medicinal plant Panax ginseng. Exp Gerontol. 1977;12:125–31.
Wang HP, Zhang YB, Yang XW, Zhao DQ, Wang YP. Rapid characterization of ginsenosides in the roots and rhizomes of Panax ginseng by UPLC-DAD-QTOF-MS/MS and simultaneous determination of 19 ginsenosides by HPLC-ESI-MS. J Ginseng Res. 2016a;40:382–94. https://doi.org/10.1016/j.jgr.2015.12.001.
Zhou SS, Xu J, Kong M, Yip KM, Di Xu J, Shen H, Zhao ZZ, Li SL, Chen HB. Synchronous characterization of carbohydrates and ginsenosides yields deeper insights into the processing chemistry of ginseng. J Pharm Biomed Anal. 2017;145:59–70. https://doi.org/10.1016/j.jpba.2017.06.042.
Yang W, Qiao X, Li K, Fan J, Bo T, Guo DA, Ye M. Identification and differentiation of Panax ginseng, Panax quinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. Acta Pharm Sin B. 2016;6:568–75. https://doi.org/10.1016/j.apsb.2016.05.005.
Liu Z, Wang CZ, Zhu XY, Wan JY, Zhang J, Li W, Ruan CC, Yuan CS. Dynamic changes in neutral and acidic ginsenosides with different cultivation ages and harvest seasons: identification of chemical characteristics for Panax ginseng quality control. Molecules. 2017;22. https://doi.org/10.3390/molecules22050734.
Harnly J, Chen P, Harrington PDB. Probability of identification: adulteration of American ginseng with Asian ginseng. J AOAC Int. 2013;96:1258–65. https://doi.org/10.5740/jaoacint.13-290.
Zhang JJ, Su H, Zhang L, Liao BS, Xiao SM, Dong LL, Hu ZG, Wang P, Li XW, Huang ZH, Gao ZM, Zhang LJ, Shen L, Cheng RY, Xu J, Chen SL. Comprehensive characterization for ginsenosides biosynthesis in ginseng root by integration analysis of chemical and transcriptome. Molecules. 2017a;22. https://doi.org/10.3390/molecules22060889.
Kim JH, Kim MK, Wang H, Lee HN, Jin CG, Kwon WS, Yang DC. Discrimination of Korean ginseng (Panax ginseng meyer) cultivar Chunpoong and American ginseng (Panax quinquefolius) using the auxin repressed protein gene. J Ginseng Res. 2016;40:395–9. https://doi.org/10.1016/j.jgr.2015.12.002.
Wang H, Wang J, Li G. A simple real-time polymerase chain reaction (PCR)-based assay for authentication of the Chinese Panax ginseng cultivar Damaya from a local ginseng population. Genet Mol Res. 2016b;15:1–10. https://doi.org/10.4238/gmr.15028801.
Wang L, Zhao S-J, Cao H, Sun Y. The isolation and characterization of dammarenediol synthase gene from Panax quinquefolius and its heterologous co-expression with cytochrome P450 gene PqD12H in yeast. Funct Integr Genomics. 2014;14:545–57.
Zhou H, Wu W, Liu L, Cheng C. Method for molecular authentication of Panax ginseng and Panax quinquefolius using SNP markers in dammarenediol synthase (DS) gene. Pat Appl:AU2015100441.
Qin J, Leung FC, Fung Y, Zhu D, Lin B. Rapid authentication of ginseng species using microchip electrophoresis with laser-induced fluorescence detection. Anal Bioanal Chem. 2005;381:812–9. https://doi.org/10.1007/s00216-004-2889-2.
Koch CA, Li PCH, Utkhede RS. Evaluation of thin films of agarose on glass for hybridization of DNA to identify plant pathogens with microarray technology. Anal Biochem. 2005;342:93–102. https://doi.org/10.1016/j.ab.2005.04.010.
Sedighi A, Li PCH. Kras gene codon 12 mutation detection enabled by gold nanoparticles conducted in a nanobioarray chip. Anal Biochem. 2014;448:58–64. https://doi.org/10.1016/j.ab.2013.11.019.
Han JY, Kim HJ, Kwon YS, Choi YE. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52:2062–73. https://doi.org/10.1093/pcp/pcr150.
Wang L, Li PCH. Gold nanoparticle-assisted single base-pair mismatch discrimination on a microfluidic microarray device. Biomicrofluidics. 2010:4. https://doi.org/10.1063/1.3463720.
Sedighi A, Whitehall V, Li PCH. Enhanced destabilization of mismatched DNA using gold nanoparticles offers specificity without compromising sensitivity for nucleic acid analyses. Nano Res. 2015;8:3922–33. https://doi.org/10.1007/s12274-015-0893-9.
Wang L, Li PCH. Flexible microarray construction and fast DNA hybridization conducted on a microfluidic chip for greenhouse plant fungal pathogen detection. J Agric Food Chem. 2007;55:10509–16.
Stevens PW, Henry MR, Kelso DM. DNA hybridization on microparticles: determining capture-probe density and equilibrium dissociation constants. Nucleic Acids Res. 1999;27:1719–27. https://doi.org/10.1093/nar/27.7.1719.
Lee Y, Kim Y, Lee D, Roy D, Park JW. Quantification of fewer than ten copies of a DNA biomarker without amplification or labeling. J Am Chem Soc. 2016;138:7075–81. https://doi.org/10.1021/jacs.6b02791.
Di Iorio D, Huskens J. Surface modification with control over ligand density for the study of multivalent biological systems. ChemistryOpen. 2020;9:53–66. https://doi.org/10.1002/open.201900290.
Chim W, Sedighi A, Brown CL, Pantophlet R, Li PCH. Effect of buffer composition on PNA-RNA hybridization studied in the microfluidic microarray chip. Can J Chem. 2018;96:241–7. https://doi.org/10.1139/cjc-2017-0319.
Zheng Y, Bergold A, Duffel MW. Affinity labeling of aryl sulfotransferase IV. Identification of a peptide sequence at the binding site for 3’-phosphoadenosine-5’-phosphosulfate. J Biol Chem. 1994;269:30313–9.
Zhang J, Fang J, Duan W, Wu L, Zhang A, Dalchau N, Yordanov B, Petersen R, Phillips A, Zhang DY. Predicting DNA hybridization kinetics from sequence. Nat Chem. 2017b;10:91–8. https://doi.org/10.1101/149427.
Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15.
Wang L, Kropinski MC, Li PCH. Analysis and modeling of flow in rotating spiral microchannels: towards math-aided design of microfluidic systems using centrifugal pumping. Lab Chip. 2011;11:2097–108. https://doi.org/10.1039/c0lc00599a.
Funding
This work received funding from NSERC and Macan Biotechnologies Ltd. of Macau.
Author information
Authors and Affiliations
Contributions
Christopher Oberc contributed by designing and conducting all experiments and wrote the paper; Abootaleb Sedighi provided advice on designing experiments; Paul C.H. Li, designed the experiments, wrote and revised the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 244 kb)
Rights and permissions
About this article
Cite this article
Oberc, C., Sedighi, A. & Li, P.C.H. The genetic authentication of Panax ginseng and Panax quinquefolius based on using single nucleotide polymorphism (SNP) conducted in a nucleic acid test chip. Anal Bioanal Chem 414, 3987–3998 (2022). https://doi.org/10.1007/s00216-022-04044-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04044-0