Abstract
In recent years, we have seen a steady rise in the prevalence of antibiotic-resistant bacteria. This creates many challenges in treating patients who carry these infections, as well as stopping and preventing outbreaks. Identifying these resistant bacteria is critical for treatment decisions and epidemiological studies. However, current methods for identification of resistance either require long cultivation steps or expensive reagents. Raman spectroscopy has been shown in the past to enable the rapid identification of bacterial strains from single cells and cultures. In this study, Raman spectroscopy was applied for the differentiation of resistant and sensitive strains of Escherichia coli. Our focus was on clinical multi-resistant (extended-spectrum β-lactam and carbapenem-resistant) bacteria from hospital patients. The spectra were collected using both UV resonance Raman spectroscopy in bulk and single-cell Raman microspectroscopy, without exposure to antibiotics. We found resistant strains have a higher nucleic acid/protein ratio, and used the spectra to train a machine learning model that differentiates resistant and sensitive strains. In addition, we applied a majority of voting system to both improve the accuracy of our models and make them more applicable for a clinical setting. This method could allow rapid and accurate identification of antibiotic resistant bacteria, and thus improve public health.
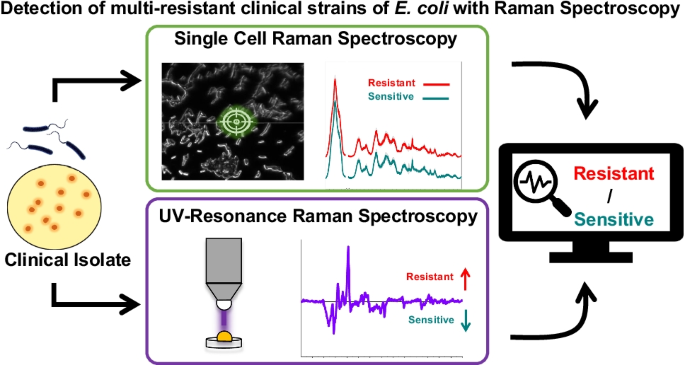
Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rise of antimicrobial resistance is a global public health challenge [1]. The Organisation for Economic Co-operation and Development (OECD) predicts that 2.4 million people in Europe, North America, and Australia will die from infections caused by resistant microorganisms before 2050, leading to healthcare costs up to US$3.5 billion per year [2, 3]. In a recent study, it was shown that ~ 28% of all resistance-attributed-deaths are caused by Escherichia coli strains that are resistant to extended-spectrum β-lactams (ESBLs) or carbapenems (CREs) [2, 4]. As these bacteria are spreading and evolving throughout the world, global actions are taken to reduce the unnecessary use of last-resort antibiotics [1, 3]. However, due to the limitations of microbiological diagnostic methods, this is often not feasible.
Prescribing the correct antibiotic treatment to patients, in time, is of paramount importance for this cause [5]. The routine microbiological techniques used in clinical laboratories require at least 48 h and up to 4 days to deliver results on pathogen resistance [6, 7]. This leads physicians to use empirical treatment based on patient history and resistance rates of healthcare facilities. These treatments are not always appropriate and contribute to further increase antimicrobial resistance, since often last-line antibiotics are used unnecessarily [5, 8].
Therefore, there is a need for rapid and reliable diagnostic procedures to facilitate prompt and effective treatment. Over the past decades, novel molecular diagnostic techniques for the classification of pathogens and detection of resistance have been developed and applied. These methods have improved clinical diagnostics significantly, but have several disadvantages, primarily the high cost of consumables and the need for extensive infrastructure and specially trained personnel [6]. Furthermore, in case of the PCR assays, the method does not apply when new mutations naturally appear as previous knowledge is needed to design the assay [9, 10]. Thus, the need for a sensitive, specific, rapid, and reliable method for antibiotic resistance detection remains.
Raman spectroscopy is a novel technique with high potential for clinical applications. It is rapiand divergent evolution. Thed and label-free and can be applied on bacterial cells with minimal sample preparation [11,12,13]. Raman spectroscopy probes the chemical fingerprint of bacterial cells and cultures, without the need for external reagents and expensive consumables. Since different strains of bacteria differ in their chemical composition, these differences are captured in their Raman spectrum and can be evaluated using chemometric methods [12, 14].
This technique has been shown in many studies to enable differentiation of bacterial strains and species [14,15,16,17,18,19,20,21,22,23,24,25]. For example, Raman microspectroscopy of single bacterial cells was used to differentiate strains of Mycobacteria [17], Burkholderia [16, 26], Legionella [19], Escherichia [15], and other clinically relevant bacteria [11, 27]. Alternatively, instead of measuring the spectra of single cells, one can also measure colonies or dried biomass. Raman spectroscopy on bulk samples was used to differentiate strains of Staphylococci [21, 23, 25], Pseudomonas [24], and other clinically relevant bacteria [20, 22].
Bulk measurement has advantages and disadvantages over single-cell analysis: it requires longer cultivation or enrichment, which takes time, but often produces better signal-to-noise ratios (S/N). This improvement is because the signal is coming from thousands of cells at a time. Moreover, in single-cell microspectroscopy, it is not practical to use ultraviolet (UV) illumination as the cell is destroyed from the high energy of the beam. UV light causes photothermal damage from the accumulated thermal energy applied to the sample, causing complex molecular structures to break down and burn. For bulk samples, this problem is mediated by rotating the slide containing the bacterial biomass in a spiral manner during measurement, minimizing local thermal build up. However, in single-cell analysis, the beam must be focused on the cell, destroying it entirely via photothermal damage.
Using UV light is important as it causes a resonance Raman signal enhancement of nucleic acids and aromatic amino acids—which improves S/N in spectra of cells tremendously [28,29,30]. When using UVRR spectroscopy on microbiological samples, the signals collected are primarily those that are resonance-enhanced—i.e., proteins, DNA, and RNA. On the other hand, if the samples are illuminated with 532 nm light, a more comprehensive look into the biochemistry of the cell is observed, with spectral features of lipids, carbohydrates proteins, and nucleic acids combined. This trade-off between higher S/N ratio and the number of spectral features observable often makes the choice of method for analysis difficult, yet, for difficult classification tasks, UVRR spectroscopy is often preferred [28]. While both single-cell analysis and bulk measurement have advantages and disadvantages important in adjusting Raman-based diagnostics to healthcare, no study so far has compared these two approaches for these applications.
Antibiotic-resistant bacteria have also been analyzed with Raman spectroscopy. For example, by exposing cells to antibiotics, resistant bacteria displayed an increase in a protein marker band and a decrease in a nucleic acid marker [31,32,33]. Some studies focused on differentiation between strains of antibiotic-resistant bacteria without antibiotic exposure. Namely, methicillin-resistant Staphylococcus aureus [23] and multi-resistant E. coli [22, 23] were typed using Raman spectroscopy on bulk samples. In those studies, it was concluded that Raman typing can be used to determine whether different clinical isolates are of the same clonal line. Despite the fact that these studies could distinguish the different strains, their approach leads to a purely taxonomic evaluation that is not suitable for everyday clinical laboratory practice. This limits the application of the method to the strains used in the study and does not consider the natural differences that define their resistances. The differences between resistant strains are many and complex, coming from diverse origins and divergent evolution. The resistance genes carried by a strain are, among others, part of the supra genome that contains plasmids, mobile genetic elements, and the bacterial chromosome. This supra genome significantly varies from strain to strain [34,35,36,37,38]. These variations are present even when the strains evolve in similar environments, such as a hospital. Another approach was to use artificially engineered antibiotic-resistant E. coli [39]. However, artificial selection for antimicrobial resistance creates strains with less diversity than those which evolve naturally and independently in natural and clinical environments [38].
The aim of the present study was to differentiate clinical multi-resistant (ESBL and CRE) E. coli isolates from sensitive strains without exposure to antibiotics. Furthermore, for the first time, the use of Raman microspectroscopy on single cells and UV resonance Raman spectroscopy (UVRR) on bulk samples was compared for the purpose of differentiation between multi-resistant and sensitive bacteria. These findings could be applied to reduce the time, cost, and effort needed to detect antibiotic resistance in clinical settings.
Material and methods
Antibiotic susceptibility test
Antibiotic susceptibility to extended-spectrum β-lactam and carbapenem antibiotics was evaluated using a VITEK 2 Compact (bioMérieux) with VITEK® 2 AST cards (bioMérieux). The test was run according to the manufacturer’s instructions. Furthermore, for isolates defined as “pathogenic/sensitive” a minimum inhibitory concentration (MIC) test was conducted for the following antibiotics: tetracycline, streptomycin, polymyxin B, gentamycin, nitrofurantoin, chloramphenicol, trimethoprim, cefuroxime, cefaclor, cefazolin, and amikacin. The tests were conducted using the E-Test (Liofilchem) according to the manufacturer’s instruction. Briefly, for each strain, 0.5 McFarland was spread on Mueller Hinton agar (Merck, Darmstadt, Germany) plates and exposed to a strip of varying antibiotic concentrations. The plates were incubated for 24 h in 37 °C, and the zone of growth inhibition was assessed visually to determine the MIC.
Sample preparation
The resistance class and sources of all E. coli used isolates are shown in Table S1. A total of 20 isolates were used, consisting of 10 multi-resistant (5 ESBL and 5 CRE) and 10 sensitive E. coli isolates. On every measurement date, bacteria were transferred from – 80 °C storage to a nutrient agar (NA) plate (Carl Roth) and incubated overnight at 37 °C. A loopful of biomass was transferred from the agar plate to Nutrient Broth media (Carl Roth) and incubated at 37 °C with shaking of 120 rpm. For each isolate, three independent replicates were prepared and measured on different days.
For UVRR measurements, the bacteria were harvested after 1 h of growth in 20 mL of Nutrient Broth media for heat inactivation. Three separate tubes of 1.5 mL inoculum were heat-inactivated at 99 °C for 5 min. The heat-inactivated bacteria were washed 3 times with 1 mL distilled water. After centrifugation at 5000 g for 5 min (Rotina 380R, Hettich), the supernatant was discarded and the pellet re-suspended in 30 μL of distilled water. Finally, bacteria were loaded onto a fused-silica slide and air dried at room temperature for 1 h. Heat inactivation was verified by plating the bacteria on NA agar plates and observing no colonies after incubation of 24 h in 37 °C.
For Raman microspectroscopy, the bacteria were grown overnight (16–24 h) in 5 mL of Nutrient Broth media. The bacteria were then washed 3 times as described above. Finally, 10 μL of the washed bacterial cell suspension was put in droplets on Ni-foil discs and allowed to dry at room temperature for 15–60 min.
Raman measurements
UVRR spectra were collected using a Raman microscope (HR800, Horiba/Jobin–Yvon) with a focal length of 800 mm. 244 nm of light was used for excitation, produced by a frequency-doubled line of an argon-ion laser (Coherent Innova 300, FReD). The laser was focused and directed through a × 40 antireflection-coated objective (LMU, NA: 0.5, UVB). Backscattered Raman light was collected through a 400-μm entrance slit into a 2400-lines/mm grating and detected by a nitrogen-cooled CCD camera. The spectral resolution was 2 cm−1. For each spectrum measured, 15 s of illumination time, and a maximal laser power of about 18 mW, was used leading to about 0.5 mW on the sample. During measurement, in order to avoid burning of the sample, the sample stage was rotated constantly in spiral manner. In each measurement, a time series of 10 spectra was obtained. A total of 25 time series were measured for each strain and replicate from 3 fused-silica slides. The spectra of each time series were averaged and considered as one spectrum to reduce noise. A total of ~ 750 spectra for each group (750 for the resistant group and 748 for the sensitive) were analyzed.
To measure individual bacterial cells using Raman microspectroscopy, a Raman microscope BioParticleExplorer (MicrobioID 0.5, RapID) was used. A 532-nm frequency-doubled solid-state Nd:YAG diode pumped laser (LCM-S-111, Laser-Export Company Ltd.) was used for excitation. The laser beam was focused with a × 100 magnification objective (MPLFLN × 100, NA: 0.9, Olympus Corporation) on the sample with a laser power of approximately 16 mW, leading to approximately 3.5 mW on the cells. Backscattered Raman light was focused to a single-stage monochromator (HE 532, Horiba Jobin Yvon) equipped with a 920-lines/mm grating and collected with a thermoelectrically cooled CCD camera (DV401A-BV, Andor Technology). The spectral resolution was approximately 10 cm−1. For each bacterial cell, two consecutive Raman spectra were measured at the same position, which were afterward combined. Integration time was 15 s for each bacterial cell. For each replicate, 60 spectra were collected. A total of > 2000 spectra for each group (2106 for the resistant group and 2062 for the sensitive) were analyzed.
Statistical analysis
Prior to analysis, several preprocessing steps were conducted using the Ramanmetrix software [40]. Preprocessing included de-spiking the spectra (based on a method described before [41]), wavenumber calibration, background correction with a sensitive nonlinear iterative peak (SNIP) algorithm using 40 iterations, vector normalization, and cutting the spectra to the relevant range (500–1900 cm−1 for UVRR spectra and 400–3050 cm−1 for Raman microspectroscopy data). Also, for the Raman microspectroscopy data the silent region (1800–2800 cm−1) was removed.
Wavenumber calibration, with a polynomial fit function, was based on polystyrene spectra and 4-acetamidophenol spectra for UVRR and Raman microspectroscopy data, respectively. The polynomial degree was 2 for the UVRR spectra and 3 for the Raman microspectroscopy. A new reference spectrum was used for every measurement date.
Classification models were calculated individually for each dataset (UVRR and Raman microspectroscopy) using the Ramanmetrix software. We compared principal component analysis support vector machine (PCA-SVM) and principal component analysis linear discriminant analysis (PCA-LDA) models for both datasets and chose the optimal model and number of principal components based on the results of leave-one-strain-out cross-validation (LOSOCV). In this validation method, a model is trained repeatedly on the dataset excluding one strain, which is then predicted by the constructed model.
For the UVRR data, a PCA-SVM model was calculated, based on 4 PCs. For the Raman microspectroscopy data, a PCA-LDA model was calculated, based on 14 PCs.
For the Raman microspectroscopy data, prior to analysis, burned spectra were removed automatically using an in-house R script [42]. Furthermore, outliers were removed from the single-cell Raman spectra by using a correlation filter. In this method, the Pearson correlation coefficient with the average preprocessed spectrum of the entire dataset is calculated for each spectrum as described before [43]. The spectra with correlation values below 0.9 were excluded from the further analysis. More than 98% of spectra passed the filter.
After the classification models were calculated, we applied a “Majority Voting” approach. Because of in-sample heterogeneity, some spectra will classify incorrectly. In order to reach a decision for a strain (resistant/sensitive), a “vote” is conducted, and the class which has the majority of spectra is chosen as the “decided class.” This was accomplished using an in-house R script.
Results and discussion
Comparison of Raman spectra of resistant and sensitive bacteria
We measured the Raman spectra of 20 E. coli strains with two spectroscopic approaches: UV resonance Raman (UVRR) spectroscopy with excitation wavelength of 244 nm and Raman microspectroscopy with excitation wavelength of 532 nm. The mean spectra, comparing the resistant and sensitive strains are presented in Fig. 1. Figure 1A presents the results of UVRR spectroscopy and Fig. 1B the results of Raman microspectroscopy. The standard deviation of each mean spectrum is highlighted in gray.
The spectra measured by UVRR on bulk samples (Fig. 1A) represent primarily bands which are enhanced by the resonance effect of excitation with 244 nm. We selected this wavelength because of the resonance enhancement of the Raman signal. This enhancement occurs for molecules with absorption bands that are near the spectral region of the laser excitation. When excited with 244 nm light, the side chain vibrations of aromatic amino acids as well as the vibrations of the nucleobases of nucleic acids are enhanced. These represent two critical components of a bacterial cell: proteins and nucleic acids [28].
The spectrum exhibits a well-established pattern that is typical for bacterial spectra, where the peaks at 786, 1242, 1335, 1362, 1485, 1530, and 1575 cm−1 originate primarily from nucleic acid residues and the peaks at 762, 831, 855, 1014, 1176, 1212, and 1617 cm−1 from aromatic amino acids [28, 39, 44,45,46,47,48]. The peaks at 1242 and 1335 cm−1 represent a mixture of signals from nucleic acids and aromatic amino acids [44]. The exact band assignment can be seen in Table S3.
In Fig. 1B, the spectra collected from single cells with Raman microspectroscopy are presented. Here, as the samples were illuminated with 532 nm light, no strong resonance effect is seen. Also, the spectra exhibit a well-established pattern, typical for bacterial spectra, which represents a large array of biomolecules [11, 12, 14]. The peaks at 2933, 1448, and 854 cm−1 represent C-H stretching vibrations, CH2 deformation vibrations, and C–C/C–O–C stretching vibrations, respectively, which are abundant in many lipid, protein, and carbohydrate molecules. The peaks at 1667 and 1241 cm−1 are from amide I and amide III vibrations, respectively, and together with the sharp peak of the phenylalanine ring breathing vibration (1004 cm−1) represent proteins in the cell. The wide peak at 1331 cm−1 represents CH2 deformation vibrations in proteins and ring vibrations of guanine and adenine [49]. Lastly, weaker bands, such as 1574, 1103, 782, and 724 cm−1, represent nucleic acids [50, 51]. The full band assignment is presented in Table S4. Taken all together, these features present a larger, more phenotypic outlook on the chemistry of the cell than the one obtained by UVRR spectroscopy.
For both approaches, no clear difference can be discerned between the groups by observing the mean spectra. This finding is in accordance with previous works on the application of Raman spectroscopy on bacterial samples [11, 12, 14, 17, 28]. The chemical fingerprint of all bacterial species includes the same biomolecules (mostly nucleic acids, proteins, and lipids). As these differ slightly in proportions, structures, and composition, only slight differences exist between the spectra of different bacteria. The closer two strains are related, the smaller these differences are. This is especially evident here as all spectra examined come from the same species of bacteria, grown in the same conditions.
However, slight differences can be found by multivariate statistical analysis and by calculating a difference spectrum. A difference spectrum was calculated by subtracting the mean spectrum of sensitive strains from that of resistant ones. In Fig. 2, difference spectra were calculated by subtracting the mean spectrum of sensitive strains from the mean spectrum of resistant ones: (A) spectrum derived from UVRR spectroscopy and (B) spectrum derived from Raman microspectroscopy. The spectra are normalized and are presented in the same scale. The difference spectrum is presented for both UVRR and Raman microspectroscopy, and the peak assignments are detailed in Fig. 2. For the UVRR spectrum, as detailed in Table 1, the Raman bands which have a stronger signal in resistant bacteria are 1578 and 1536 (guanine), 1488 (nucleic acid marker), and 789 cm−1 (cytosine) [39, 52, 53]. All these vibrations represent nucleic acid residues. In contrast, the Raman signals which are enhanced for sensitive strains were 1659, 1614, 1206, 1176, 1005, and 852 cm−1. These peaks represent the aromatic amino acids phenylalanine, tyrosine, tryptophan, and histidine, which are indicators for proteins [28]. These findings suggest that in resistant bacteria, we see an increase in nucleic acid/protein ratio.
For the Raman microspectroscopy data, where no resonance effect is present, bands of all biomolecules are present in both the resistant and sensitive isolates. The assignments are given in Table 2. Raman bands which have a stronger signal in resistant bacteria include nucleic acids, represented by cytosine and thymine bands at 1640 and 1361 cm−1 and proteins, represented by bands 1064 and 1004 cm−1 (C–C/C-N stretching vibrations and the ring breathing of phenylalanine, respectively). Lastly, the bands assigned to lipids are at 884 cm−1 (C-O–O skeletal vibrations) and 1451 cm−1 (CH2 deformation vibrations). Whereas for sensitive isolates, the nucleic acids are represented by vibrations at 728, 1484, and 1577 cm−1 (guanine, adenine), proteins at 635 (tryosine skeletal vibrations), and lastly lipids by the band at 2849 cm−1 (for CH2 stretching vibrations). These features show again the presence of a phenotypic outlook of the bacterial biochemical composition that is detected by the 532 nm excitation. However, the difference spectrum consists of broad, superimposed Raman signals with low intensity. Although there are peaks identifiable, they are not as prominent as in UVRR and are therefore difficult to interpret and negligible (Fig. 2B).
The findings from the UVRR difference spectrum can be explained by the fact that multi-resistant strains carry, beside their core genome, resistance genes. These genes are present on plasmids, bacterial chromosomes, and mobile genetic elements. When translated, they allow the bacteria to overcome the toxic effect of antibiotics in their environment. Resistance genes, which developed with the extensive use of antibiotics over the past 80 years, are part of an evolutionary process in the genome of multi-resistant strains [57]. These genes affect the composition of DNA in the cell both qualitatively and quantitatively: as resistant strains contain more genetic material compared to sensitive ones. This change in the genetic composition is likely the cause of the differences seen with UVRR spectroscopy.
In the 532 nm excitation, no resonance enhancement of the nucleic acids occurs, and thus, the small changes in nucleic acid content and composition are not observed. From a biochemical perspective, the overall metabolism of these different strains does not appear to have changed significantly. This is not surprising, since the bacteria were not exposed to antibiotics. Resistance mechanisms are often not expressed unless the cell is under stress and therefore the metabolism and phenotype remained nearly identical.
Recently, a similar Raman setup was used to study the difference between sensitive strains of E. coli and strains which were engineered to be resistant. One study simulated the acquisition of a resistance plasmid in a native E. coli strain by molecular engineering, and measured the bacteria with Raman microspectroscopy and UV resonance Raman spectroscopy [39]. Another study used Raman spectroscopy to discriminate between lab-evolved resistant E. coli strains [58]. Both studies found that the differentiation between the resistant and sensitive strains was indeed related to the nucleic acid/protein ratios. Moreover, in both studies, the ratio was larger for resistant strains. This supports our similar findings in the present study on clinical, naturally occurring bacteria.
Classification of resistant and sensitive strains
In order to assess the ability of Raman spectroscopy to differentiate between resistant and sensitive strains, a machine learning classification model was trained. Two common classification models, SVM for UVRR and LDA for Raman microspectroscopy, were used to analyze the differences in the spectra. The model parameters, such as number of principal components, were optimized using the leave-one-strain-out cross-validation (LOSOCV) method. The classification results are presented in Table 3. In each table, Raman spectra were classified into their respective class. The accuracy, sensitivity, and specificity of each model are also presented. For both UVRR data and Raman microspectroscopy data, the models performed with 60% accuracy. This indicates that the differentiation of resistant and sensitive strains is possible, though limited, with both methods by applying machine learning algorithms.
This finding is surprising given that the difference spectra of the two methods differed significantly. However, this antagonism can be explained by the dimension reduction and manipulation of the data by the machine learning method. These methods highlight slight differences and revalues common features. Thus, even very minor differences, which are not observable in the spectra can be of significance, as demonstrated before [11, 59].
Application of majority voting to improve classification
When measuring bacteria with Raman spectroscopy, many spectra are measured for each isolate. Since not all the bacteria are perfectly identical, a natural heterogeneity can be seen in the spectra. Especially in the single cell measurements the heterogeneity is larger due to cell to cell variations originating from differences in growth stage and metabolism. In bulk measurements, heterogeneity is not as intense, since each spectrum is composed of signals from thousands of cells. For clinical decision-making, there is a need for precise results, with concrete information about the resistance characteristics. Therefore, this heterogeneity must be resolved, providing a read-out that does not refer to individual spectra but to an entire bacterial strain.
To eliminate this problem, we applied a majority voting system on the confusion matrix of the classification models. In this system, the sum of decisions is calculated and a vote is taken, where for each strain, a majority of classifications decides whether the strain is classified as resistant or sensitive. The result of majority voting per strain is shown in Table 4. For the UVRR spectroscopy data, the accuracy of the model improved from 60% in the original model to 70% after voting, reaching a correct classification of 14/20 strains. For the Raman microspectroscopy data, we observed an increase in accuracy, from 60 to 75%, correctly classifying 15/20 strains.
This means that overall accuracy, with majority voting, has increased by 10–15%. For the UVRR data, the sensitivity of the model improves much more, from 60 to 90% (meaning 9/10 resistant strains were correctly identified). Furthermore, the problem of sample heterogeneity has been mediated; as for each strain, only one decision remains. The increase in accuracy, together with removal of heterogeneity, helps define the presence of resistance in the strains and is useful for clinical decision-making.
The misclassified strains are summarized in Table S5. Interestingly, only the sensitive strain DSM 498 is misclassified by both methods. The other nine misclassified strains are unique.
Overall, we found that UVRR spectroscopy and Raman microspectroscopy provide similar classification results. This finding is surprising because we expected the UVRR to significantly outperform Raman microspectroscopy due to the stronger S/R ratio and the differences in the spectra (Fig. 2). The ability to differentiate resistance from UVRR was mostly dependent on resonance-enhanced signals. For single-cell spectra excited with 532 nm light, this resonance enhancement does not occur. It is likely that for these spectra, the classification is related not only to changes in genetic content, but also to the larger metabolic changes which a resistant cell undergoes. Resistant bacteria show a decline in overall fitness even when grown in optimal conditions. This decline is related to a loss in enzyme efficiency, changes in cell wall thickness, and membrane porin content. While each of these changes is important in resistance, they will not affect the spectra dramatically. They are slight changes in the overall metabolic profile of a cell [60,61,62]. However, it seems that even such slight changes are present in the spectra, and by using a machine learning algorithm, they enabled classification as efficient as that achieved with UVRR spectroscopy. This finding is the most important finding of this study. It highlights the applicability of both approaches in detecting microbial susceptibility and can guide future studies in the flexibility of Raman-based diagnostics. With machine learning algorithms, one can extract information from nonresonant spectra that is as useful as resonance-enhanced spectra—for a clinical application.
It is important to note that since different wavelengths were employed as excitation sources, as well as different sample preparation methods (bulk and single cells), a direct comparison of the performances of UVRR spectroscopy and Raman microspectroscopy cannot be made. The same exact cell was never measured twice, and the same exact sample was not prepared for both devices. However, this is not the focus of this study. This study focuses on presenting the ability of different approaches using Raman spectroscopy and analyzing their potential individual diagnostic value.
One may argue about whether this method is suitable for clinical laboratory settings since the accuracy provided is only fair. However, the improvement of overall accuracy and sensitivity by majority voting evidenced that by removing the natural heterogeneity within the strain, the presence of resistance can be revealed. Since the number of stains used in this study was small, it is interesting to see how the diagnostic potential is shown. Further studies are required, with larger bacterial cohorts and a larger biodiversity of strains and species in order create a database suitable for hospital settings. The biggest advantage of Raman spectroscopy compared to traditional microbiology methods is its high speed in providing results. By providing treating physicians with information on bacterial resistance early, many unnecessary prescriptions of last-line antibiotics can be avoided. This could allow better management of hospital resistance rates, and hinder the rise of antimicrobial resistance.
Conclusions
In the present study for the first time, ESBL and CRE clinical E. coli isolates could be distinguished from sensitive strains using Raman spectroscopy. We show that UVRR spectra provide better S/R ratio and a change in nucleic acid/protein ratio can be observed. Yet, in contrast to what was expected, when using machine learning algorithms to classify the data, both methods were comparable. This comparison was done for the first time on clinically relevant data and future studies should consider these methods comparable to some extent. Lastly, we found majority voting is key to minimizing the natural heterogeneity and make Raman spectroscopy more suitable for clinical application.
This study presents another facet of Raman spectroscopy–based diagnostics. We conclude this method could enable better, rapid, label-free antibiotic resistance diagnostics in clinical settings. For future studies, especially those with clinical applications, we recommend applying a majority voting approach to clarify results into actionable information. Further work is still essential to improve classification for use in healthcare. The classification accuracy found in the study is limited for application, but likely with a larger and more diverse dataset, it could improve, providing better diagnostics to help fight the rise of antimicrobial resistance and for the improvement of global health.
Availability of data and material
The data presented in this study are available on request from the corresponding author.
Code availability
Not applicable.
References
World Health Organization (WHO). Global action plan on antimicrobial resistance. 2015. https://www.who.int/publications/i/item/9789241509763. Accessed 27 SEP 2021.
Hofer U. The cost of antimicrobial resistance. Nat Rev Microbiol. 2019. https://doi.org/10.1038/s41579-018-0125-x.
Organisation for Economic Co-operation and Development (OECD). Stemming the Superbug Tide. 2018. https://doi.org/10.1787/9789264307599-en.
Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019. https://doi.org/10.1016/S1473-3099(18)30605-4.
Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program*. Crit Care Med. 2014;42(8):1749–55.
AlMasoud N, Muhamadali H, Chisanga M, AlRabiah H, Lima CA, Goodacre R. Discrimination of bacteria using whole organism fingerprinting: the utility of modern physicochemical techniques for bacterial typing. Analyst. 2021;146(3):770–88.
Peacock SJ, Parkhill J, Brown NM. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology. 2018;164(10):1213–9.
Hecker MT, Aron DC, Patel NP, Lehmann MK, Donskey CJ. Unnecessary use of antimicrobials in hospitalized patients: current patterns of misuse with an emphasis on the antianaerobic spectrum of activity. Arch Intern Med. 2003;163(8):972–8.
Syal K, Mo M, Yu H, Iriya R, Jing W, Guodong S, et al. Current and emerging techniques for antibiotic susceptibility tests. Theranostics. 2017;7(7):1795–805.
Lutgring JD, Limbago BM. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J Clin Microbiol. 2016;54(3):529–34.
Lorenz B, Wichmann C, Stöckel S, Rösch P, Popp J. Cultivation-free Raman spectroscopic investigations of bacteria. Trends Microbiol. 2017;25(5):413–24.
Stöckel S, Kirchhoff J, Neugebauer U, Rösch P, Popp J. The application of Raman spectroscopy for the detection and identification of microorganisms. J Raman Spectrosc. 2015;47:89–109.
Pahlow S, Weber K, Popp J, Wood BR, Kochan K, Rüther A, et al. Application of vibrational spectroscopy and imaging to point-of-care medicine: a review. Appl Spectrosc. 2018;72(1_suppl):52–84.
Pahlow S, Meisel S, Cialla-May D, Weber K, Rösch P, Popp J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev. 2015;89:105–20.
Lorenz B, Ali N, Bocklitz T, Rösch P, Popp J. Discrimination between pathogenic and non-pathogenic E. coli strains by means of Raman microspectroscopy. Anal Bioanal Chem. 2020;412(30):8241–7. https://doi.org/10.1007/s00216-020-02957-2.
Moawad AA, Silge A, Bocklitz T, Fischer K, Rösch P, Roesler U, et al. A machine learning-based Raman spectroscopic assay for the identification of Burkholderia mallei and related species. Molecules. 2019;24(24):4516.
Stöckel S, Meisel S, Lorenz B, Kloß S, Henk S, Dees S, et al. Raman spectroscopic identification of Mycobacterium tuberculosis. J Biophotonics. 2017;10(5):727–34.
Pahlow S, Stöckel S, Pollok S, Cialla-May D, Rösch P, Weber K, et al. Rapid identification of Pseudomonas spp. via Raman spectroscopy using pyoverdine as capture probe. Anal Chem. 2016;88(3):1570–7.
Kusić D, Rösch P, Popp J. Fast label-free detection of Legionella spp. in biofilms by applying immunomagnetic beads and Raman spectroscopy. Syst Appl Microbiol. 2016;39(2):132–40.
Ho CS, Jean N, Hogan CA, Blackmon L, Jeffrey SS, Holodniy M, et al. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat Commun. 2019;10(1):4927.
Rebrošová K, Šiler M, Samek O, Růžička F, Bernatová S, Holá V, et al. Rapid identification of staphylococci by Raman spectroscopy. Sci Rep. 2017;7(1):14846.
Willemse-Erix D, Bakker-Schut T, Slagboom-Bax F, Jachtenberg J-w, Lemmens-den Toom N, Papagiannitsis CC, et al. Rapid typing of extended-spectrum β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates by use of SpectraCell RA. J Clin Microbiol. 2012;50(4):1370–5.
Wulf MWH, Willemse-Erix D, Verduin CM, Puppels G, van Belkum A, Maquelin K. The use of Raman spectroscopy in the epidemiology of methicillin-resistant Staphylococcus aureus of human- and animal-related clonal lineages. Clin Microbiol Infect. 2012;18(2):147–52.
Willemse-Erix DF, Jachtenberg JW, Schut TB, van Leeuwen W, van Belkum A, Puppels G, et al. Towards Raman-based epidemiological typing of Pseudomonas aeruginosa. J Biophotonics. 2010;3(8–9):506–11.
Willemse-Erix DF, Scholtes-Timmerman MJ, Jachtenberg JW, van Leeuwen WB, Horst-Kreft D, Bakker Schut TC, et al. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J Clin Microbiol. 2009;47(3):652–9.
Stöckel S, Meisel S, Elschner M, Melzer F, Rösch P, Popp J. Raman spectroscopic detection and identification of Burkholderia mallei and Burkholderia pseudomallei in feedstuff. Anal Bioanal Chem. 2015;407(3):787–94.
Lorenz B, Rösch P, Popp J. Isolation matters-processing blood for Raman microspectroscopic identification of bacteria. Anal Bioanal Chem. 2019;411(21):5445–54.
Silge A, Heinke R, Bocklitz T, Wiegand C, Hipler U-C, Rösch P, et al. The application of UV resonance Raman spectroscopy for the differentiation of clinically relevant Candida species. Anal Bioanal Chem. 2018;410(23):5839–47.
Asher SA. UV resonance Raman spectroscopy for analytical, physical, and biophysical chemistry. Anal Chem. 1993;65(2):59A-66A.
Gaus K, Rösch P, Petry R, Peschke KD, Ronneberger O, Burkhardt H, et al. Classification of lactic acid bacteria with UV-resonance Raman spectroscopy. Biopolymers. 2006;82(4):286–90.
Kirchhoff J, Glaser U, Bohnert JA, Pletz MW, Popp J, Neugebauer U. Simple ciprofloxacin resistance test and determination of minimal inhibitory concentration within 2 h using Raman spectroscopy. Anal Chem. 2018;90(3):1811–8.
Schröder U-C, Kirchhoff J, Hübner U, Mayer G, Glaser U, Henkel T, et al. On-chip spectroscopic assessment of microbial susceptibility to antibiotics within 3.5 hours. J Biophotonics. 2017;10(11):1547–57.
Assmann C, Kirchhoff J, Beleites C, Hey J, Kostudis S, Pfister W, et al. Identification of vancomycin interaction with Enterococcus faecalis within 30 min of interaction time using Raman spectroscopy. Anal Bioanal Chem. 2015;407(27):8343–52.
Innamorati KA, Earl JP, Aggarwal SD, Ehrlich GD, Hiller NL. The bacterial guide to designing a diversified gene portfolio. In: Tettelin H, Medini D, editors. The pangenome: diversity, dynamics and evolution of genomes. Cham: Springer International Publishing; 2020. p. 51–87.
Spencer CCA, Deloukas P, Hunt S, Mullikin J, Myers S, Silverman B, et al. The influence of recombination on human genetic diversity. PLOS Genet. 2006;2(9):e148.
Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–72.
Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007;2(5):501–12.
Klümper U, Recker M, Zhang L, Yin X, Zhang T, Buckling A, et al. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. 2019;13(12):2927–37.
Walter A, Reinicke M, Bocklitz T, Schumacher W, Rösch P, Kothe E, et al. Raman spectroscopic detection of physiology changes in plasmid-bearing Escherichia coli with and without antibiotic treatment. Anal Bioanal Chem. 2011;400(9):2763–73.
RAMANMETRIX - Online software platform for Raman spectroscopic data analysis. https://ramanmetrix.eu/. Accessed 23June 2020
Ryabchykov O, Bocklitz T, Ramoji A, Neugebauer U, Foerster M, Kroegel C, et al. Automatization of spike correction in Raman spectra of biological samples. Chemom Intell Lab Syst. 2016;155:1–6.
Team RC. R: A language and environment for statistical computing. R version 3.5.3 ed. Vienna, Austria: R Foundation for Statistical Computing; 2020.
Espagnon I, Ostrovskii D, Mathey R, Dupoy M, Joly P, Novelli-Rousseau A, et al. Direct identification of clinically relevant bacterial and yeast microcolonies and macrocolonies on solid culture media by Raman spectroscopy. J Biomed Optics. 2014;19(2):027004.
De Gelder J, De Gussem K, Vandenabeele P, Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectrosc. 2007;38(9):1133–47.
Harz M, Claus RA, Bockmeyer CL, Baum M, Rösch P, Kentouche K, et al. UV-resonance Raman spectroscopic study of human plasma of healthy donors and patients with thrombotic microangiopathy. Biopolymers. 2006;82(4):317–24.
Töpfer N, Müller MM, Dahms M, Ramoji A, Popp J, Slevogt H, et al. Raman spectroscopy reveals LPS-induced changes of biomolecular composition in monocytic THP-1 cells in a label-free manner. Integr Biol (Camb). 2019;11(3):87–98.
Zukovskaja O, Ryabchykov O, Strassburger M, Heinekamp T, Brakhage AA, Hennings JC, et al. Towards Raman spectroscopy of urine as screening tool. J Biophotonics. 2019;13(1):e201900143.
Žukovskaja O, Kloß S, Blango MG, Ryabchykov O, Kniemeyer O, Brakhage AA, et al. UV-Raman spectroscopic identification of fungal spores important for respiratory diseases. Anal Chem. 2018;90(15):8912–8.
Kloß S, Kampe B, Sachse S, Rösch P, Straube E, Pfister W, Kiehntopf M, Popp J. Culture independent Raman spectroscopic identification of urinary tract infection pathogens: a proof of principle study. Anal Chem. 2013;85:9610–6.
Maquelin K, Kirschner C, Choo-Smith LP, van den Braak N, Endtz HP, Naumann D, et al. Identification of medically relevant microorganisms by vibrational spectroscopy. J Microbiol Methods. 2002;51(3):255–71.
Huang WE, Li M, Jarvis RM, Goodacre R, Banwart SA. Chapter 5 - shining light on the microbial world: the application of Raman microspectroscopy. Adv Appl Microbiol. Academic Press. 2010;70:153–86.
Schuster KC, Urlaub E, Gapes JR. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J Microbiol Methods. 2000;42(1):29–38.
Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, et al. Nonresonant Raman imaging of protein distribution in single human cells. Biopolymers. 2003;72(1):1–9.
Tu AT. Raman spectroscopy in biology: principles and applications. New York: Wiley; 1982.
Azemtsop Matanfack G, Taubert M, Guo S, Bocklitz T, Küsel K, Rösch P, et al. Monitoring deuterium uptake in single bacterial cells via two-dimensional Raman correlation spectroscopy. Anal Chem. 2021;93(21):7714–23.
Czamara K, Majzner K, Pacia MZ, Kochan K, Kaczor A, Baranska M. Raman spectroscopy of lipids: a review. J Raman Spectrosc. 2015;46(1):4–20.
De Angelis G, Del Giacomo P, Posteraro B, Sanguinetti M, Tumbarello M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int J Mol Sci. 2020;21(14):5090.
Germond A, Ichimura T, Horinouchi T, Fujita H, Furusawa C, Watanabe TM. Raman spectral signature reflects transcriptomic features of antibiotic resistance in Escherichia coli. Commun Biol. 2018;1:85.
Ali N, Girnus S, Rösch P, Popp J, Bocklitz T. Sample-size planning for multivariate data: a Raman-spectroscopy-based example. Anal Chem. 2018;90(21):12485–92.
Stapleton Paul D, Shannon Kevin P, French GL. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 β-lactamase production and loss of an outer membrane protein. Antimicrob Agents Chemother. 1999;43(5):1206–10.
Little ML, Qin X, Zerr DM, Weissman SJ. Molecular diversity in mechanisms of carbapenem resistance in paediatric Enterobacteriaceae. Int J Antimicrob Agents. 2012;39(1):52–7.
Melnyk AH, Wong A, Kassen R. The fitness costs of antibiotic resistance mutations. Evol Appl. 2015;8(3):273–83.
Acknowledgements
We would like to thank Darina Storozhuk for her part in the development and support of the Ramanmetrix software. We acknowledge the contribution of Julian Hniopek in writing and troubleshooting some of the code used in this paper. Lastly, bacterial isolates were graciously provided by our collaborators from the University of Thessaly: Prof. Charalambos Billinis and Prof. Efthimia Petinaki as well as Prof. Ralf Ehricht of the Leibniz Institute of Photonic Technology, Jena, Germany.
Funding
Open Access funding enabled and organized by Projekt DEAL. Financial support of this study was provided by the German Federal Ministry of Education and Research as part of project CarbaTech (FKZ 01EI1701) and the research campus InfectoGnostics (FKZ 13GW0096F) and is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
Conceptualization, methodology, investigation, writing—original draft preparation, A.N. and A.P.; data analysis, writing—original draft preparation, O.R. and T.W.B.; writing—review and editing, P.R. and J.P. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nakar, A., Pistiki, A., Ryabchykov, O. et al. Detection of multi-resistant clinical strains of E. coli with Raman spectroscopy. Anal Bioanal Chem 414, 1481–1492 (2022). https://doi.org/10.1007/s00216-021-03800-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03800-y






