Abstract
In this study, Lba Cas12a (Cpf1) as one of the CRISPR systems from Lachnospiraceae bacterium was coupled with a hybridization chain reaction (HCR) to develop an electrochemical biosensor for detecting the pathogenic bacterium, Salmonella typhimurium. Autonomous cross-opening of functional DNA hairpin structures of HCR yielded polymer double-stranded DNA wires consisting of numerous single-stranded DNAs, which initiated the trans-cleavage activity of CRISPR-Cas12a to indiscriminately cleave random single-stranded DNA labeling electrochemical tags on the surface of the electrode. It led to a variation in the electron transfer of electrochemical tags. The polymer double-stranded DNA of HCR was immobilized on dynabeads (DBs) via the S. typhimurium aptamer and released from DBs. The established method could selectively and sensitively quantify S. typhimurium in samples with detection limits of 20 CFU/mL. Our study provides a novel insight for exploring universal analytical methods for pathogenic bacteria based on CRISPR-Cas12a coupled with HCR.
Similar content being viewed by others
Introduction
Food safety concerns caused by foodborne pathogenic bacteria have attracted increasing attention worldwide. Salmonella typhimurium is one of the major foodborne pathogenic bacteria that threatens people’s health mainly through consuming contaminated foods [1]. The existing gold-standard methods for detecting S. typhimurium mainly include culture methods, enzyme-linked immunosorbent assays (ELISA), polymerase chain reactions (PCR), and so forth. However, these methods have some disadvantages. For example, culture is time-consuming and generally needs 2–3 days. PCR often requires a complex DNA extraction procedure [2]. ELISA lacks sufficient sensitivity [3]. The methods require well-trained technicians and expensive equipment for implementation. Therefore, sensitive and simple approaches for detecting S. typhimurium need to be developed to ensure food safety.
CRISPR-Cas12a systems generally function as RNA-guided endonucleases [4]. Cas12a with its trans-activity triggered by dsDNA or ssDNA can cleave the nearby nonspecific ssDNA [5]. Based on the nonspecific cleavage of labeled nucleic acid reporters, the CRISPR-Cas12a system along with trans-activity can detect nucleic acid targets. For non-nucleic acid detection, the non-nucleic acid target was transduced into DNA or RNA for the Cas12a-gRNA binary complex. The excellent performance of CRISPR-Cas12a has gained attention in terms of developing the next-generation biosensor for diagnosis [5, 6], including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [7, 8], influenza A and B viruses [9], and African swine fever virus [10]. To detect low-abundance targets, it is often necessary to use various nucleic acid amplification strategies, such as recombinase polymerase amplification (RPA) [9, 11], loop-mediated isothermal amplification (LAMP) [12, 13], rolling circle amplification [14], and catalytic hairpin assembly [15, 16], Combining the collateral effect of Cas12a with isothermal amplification has created a rapid and specific detection platform [14].
The most widely used hybridization chain reaction (HCR) is a simple, enzyme-free, robust, and efficient isothermal amplification of nucleic acids; long, linear duplex concatamers are formed through the cross-opening and self-assembling of two DNA hairpins initiating by a target probe [7, 8]. In addition, target-free HCR amplification is a probe amplification technique; cross-contamination and false-positive results frequently occurring in LAMP, RPA, and PCR are effectively reduced [17]. To date, HCR has been explored to detect a variety of targets, such as nucleic acids, proteins, and pathogens [18]. Over the past few decades, HCR applications in biosensors have undergone significant development due to the particular use of the formed duplex concatamers in biosensors. Combining Cas12a with HCR has provided a detection method for targets [10]. Based on HCR and CRISPR-Cas12a, Xing et al. developed an aptamer-HCR-CRISPR-Cas12a fluorescence readout approach for the direct detection of tumor-derived extracellular vesicle proteins [17]. Kachwala combined HCR with CRISPR-Cas12a to develop a gel electrophoresis-based assay. The conserved ssDNA and dsDNA regions of tobacco circlet virus and hepatitis B virus genomes were detected, respectively [19]. Cas12, guided by crRNA, can recognize target dsDNA and initiate the trans-cleavage activity, but it requires protospacer-adjacent motif (PAM) sequences adjacent to the target dsDNA, which may limit its broader applications of HCR [20]. To solve these problems, the PAM sequence is introduced into the amplified products of HCR. It increases the difficulty of designing and screening two hairpins for HCR. ssDNA, as the recognizing target, activates the Cas12-crRNA complex without PAM sequences. In this study, the 3' ends of the two hairpins of HCR were labeled, and the polymer dsDNA wires formed by HCR carried multiple recognizing target sequences that activated the Cas12-crRNA complex.
In this study, we intended to introduce HCR-based CRISPR-Cas12a into an electrochemical biosensor platform for sensitive and specific detection of S. typhimurium (Scheme 1) [21] Electrochemical biosensors have been widely applied to detect pathogenic bacteria in food safety processes due to their fast signal readouts, simplicity, high sensitivity, and low cost, thus providing a practical or deployable point-of-care system [17]. The nonspecific ssDNA reporter (methylene blue (MB) tag modified on hairpin DNA (hpDNA)) was drafted with an MB electrochemical tag for signal transduction and a thiol moiety to tether on the sensor surface to acquire the signal electrically. Hence, the electron transfer process between the Au electrode and the redox-active species on the ssDNA could be electrochemically initiated and transduced [15].
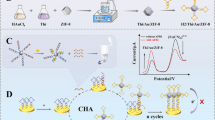
Autonomous cross-opening of functional DNA hairpin structures of HCR was used to generate a DNA double helix consisting of numerous single-stranded DNAs. These single-stranded DNAs could initiate the formation of a Cas12a-crRNA-target DNA ternary complex, which initiated the trans-cleavage activity of CRISPR-Cas12a. CRISPR-Cas12a cleaved random single-stranded DNA labeling MB electrochemical tags on the surface of the electrode (MB reporter) and led to a variation in the electron transfer of the electrochemical tag. The polymer double-stranded DNA of HCR was immobilized on DBs via the linker probe. Initially, the linker probe was locked through the S. typhimurium aptamer. Aptamers are single-stranded oligonucleotide sequences for target molecules and can efficiently and specifically bind to target molecules [22, 23]. The aptamer was shed, and HCR-binding sites were exposed. In the presence of the target, the polymer double-stranded DNAs of HCR were connected through linkers from the DBs, activated the Cas12a trans-cleavage activity, and removed the MB reporter gene from the surface of the gold electrode (GE), thereby significantly reducing the MB transduction signal. When the target was not present, no Cas12a trans-cleavage activity was observed, and the polymer double-stranded DNA of HCR was released from the DBs, thereby retaining the MB probe on the surface of the GE. The electrochemical signal output diagram based on the presence/absence of S. typhimurium is shown in Scheme 1. Based on HCR and CRISPR-Cas12a, we developed a universal electrochemical biosensor that detected pathogenic bacteria.
Experimental section
Apparatus and materials
EnGen Lba Cas12a and 10 × NE Buffer 2.1 (0.5 M NaCl, 0.1 M Tris–HCl, 0.1 M MgCl2, and 1 mg/mL BSA, pH 7.9) were provided by New England Biolabs (MA, USA). Tris (2-carboxyethyl)-phosphine hydrochloride (TCEP) and all oligonucleotides used in this study (Table S1) were purified and synthesized by Sangon Biotechnology Co. Ltd (Shanghai, China). DNase/RNase-free deionized water and the RNase inhibitor were obtained from Tiangen Biotech (Beijing) Co., Ltd. Dynabeads (DBs) M-270 Streptavidin and SYBR Safe DNA gel stain were purchased from Thermo Fisher Scientific Co., Ltd. (Norway). Furthermore, 20-bp DNA Ladder (Dye Plus) was obtained from Takara Biotech (Dalian, China). Inc. Also, 6-mercapto-1-hexanol (MCH) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid were procured from Sigma-Aldrich (St. Louis, MO, USA). The aptamer is an ssDNA that can bind to S. typhimurium with high affinity. Strains of S. typhimurium (CICC 21,484), Escherichia coli (CICC 10,389), Listeria monocytogenes (CICC 21,633), Staphylococcus aureus (CICC 21,600), and Vibrio parahaemolyticus (CICC21617) were purchased from the China Center for Type Culture Collection (Beijing, China). All reagents were diluted with DNase/RNase-free deionized water throughout the study.
Preparation of the MB probe − modified biosensing electrode
The bare Au electrode (GE) was immersed into piranha solution (H2SO4 / H2O2 = 3:1) for 20 min to remove the impurity and then scrubbed with the absorbent cotton balls soaked in deionized water. The electrode was polished with 0.05 μM alumina powder to get a smooth surface and then scrubbed with the absorbent cotton balls. The thiolated MB probe was dissolved in 1 mM phosphate-buffered saline (PBS) solution (containing 14 mM NaCl, 0.3 mM KCl, and 10 mM TCEP, pH 7.4). Then, 10 μL of 0.3 μM MB probe was added to the pretreated electrode surface and incubated overnight at room temperature. Furthermore, 10 μL of MCH (1 μM) was added dropwise onto the surface of the GE and incubated at 37 °C for 1 h. Finally, the unbound material on the surface of the electrode was removed with ultrapure water for electrochemical measurements and subsequent experiments.
Amplified ssDNA protocol by HCR
The corresponding stock solutions (100 μM) were diluted with HEPES buffer (20 mM HEPES, 8 mM MgCl2.6H2O, 20 mM NaCl, 10 mM KCL, pH 7.4) to prepare the desired concentrations of the oligonucleotides. To perform the amplified ssDNA assay, various concentrations of T were incubated with H1 (1 μM) and H2 (1 μM) in HEPES buffer (30 μL) for 1 h at 37 °C and stored at 4 °C for later use.
Target capture protocol
(1) Furthermore, 4 μL of modified streptavidin DBs was washed three times with deionized water and added to the 200-μL PCR tube. Next, the processed L-P solution (probe linker (0.5 μM) and aptamer (4 μM) mixed in equal volume) was incubated at 90 °C for 3 min, transferred immediately to an ice bath, and incubated for 60 min at 37℃. (2) The probes not bound to the DBs were washed with HEPES buffer of pH 7.4, and 50 μL of S. typhimurium and 50 μL of HEPES buffer were added for control, followed by incubation for 60 min at 37℃. (3) The DBs were washed with HEPES buffer of pH 7.4, and the pretreated HCR product was added, followed by incubation for 60 min at 37 °C. (4) The HCR-DBs were washed with HEPES buffer of pH 7.4 again, resuspended in 4 μL of HEPES buffer, and stored at 4 °C for later use.
Preparation of CRISPR-Cas12a restriction digestion system
The mixed solution containing 4 μL of Cas12a (0.3 μM), 4 μL of crRNA (1 μM), 1 μL of RNase inhibitor (10 U), and 4 μL of the HCR DBs solution resuspended was placed at room temperature for 13 min, so that Cas12a protein, crRNA, and ssDNA fragments on the HCR product were combined to enhance the activation of the CRISPR-Cas12a digestion system.
Electrochemical measurements
The electrochemical measurements were performed on the CHI660E electrochemical workstation (Shanghai Chenhua Instrument Co., Ltd., China), including electrochemical impedance spectroscopy (EIS), cyclic voltammetry (CV), and differential pulse voltammetry (DPV). The electrochemical workstation uses a classical three-electrode system in which the platinum wire is used as the auxiliary electrode, the GE is used as the working electrode, and the Ag/AgCl electrode is used as the reference electrode. The EIS and CV measurements were carried out in the buffer containing 5 mM [Fe(CN)6] 3−/4− and 0.1 M KCl. The frequency range of EIS measurement was 0.1–100 kHz, and the amplitude was 5 mV. The potential range of CV measurement was – 0.2 to 0.6 V, and the scan rate was 50 mV/s. The DPV measurement was performed in 2 × PBS solution (2.5 mM MgCl2, 50 mM NaCl, pH 7.4); the amplitude and pulse period were 50 mV and 0.5 s, respectively.
Polyacrylamide gel electrophoresis
A 12% polyacrylamide gel was used for checking the formation of HCR products using polyacrylamide gel electrophoresis (PAGE). Electrophoresis was performed in 1 × TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA, pH 8.3) at a constant voltage of 200 V for 45 min before staining with the SYBR Safe DNA gel stain. Finally, visual analysis was performed in the gel imaging system.
Sample determination
Different S. typhimurium densities (104, 105, and 106 CFU/mL) were added to sterilized milk samples for recovery analysis. HEPES buffer was used as a negative control.
Results and discussion
Characterization of HCR formation
The reaction efficiency of HCR is closely related to the sensitivity of this assay. Therefore, the PAGE assay was performed to verify the formation of HCR products (Fig. 1). The concentration of the initiator is related to the average molecular weight of the polymer produced by HCR. Too many initiators (Trigger) may cause hybridization between H1 and H2, and subsequently no adequate long chains of DNA polymers are formed. Hence, different concentrations of Trigger were used to optimize the best performance of the HCR system. As shown in Fig. 1, when the concentration of Trigger was low, there was scarcely any polymer of high bands in line 1. This finding indicated that no HCR occurred due to the lack of Trigger. The efficiency of HCR gradually improved with the increase in Trigger concentration from 0.03 to 0.3 μM, and the quantity of the hairpin probe of H1 and H2 residues decreased. When the concentration of Trigger further increased from 0.3 to 1 μM, the molecular weight of the HCR product decreased and the polymer became dispersed. This might not be conducive to effective signal amplification. Thus, the initiators of the Trigger concentration of 0.3 μM were determined to be used for the following experiments.
Electrochemical characterization of the biosensor
EIS and CV were measured using 5 mM [Fe(CN)6] 3−/4− solution as the electrochemical probe containing 100 mM KCl to investigate the construction of the biosensor. As shown in Fig. 2 and Table S2, responses of the charge transfer resistance (Rct) had a prominent increase for GE (curve a, Rct = 200.7 Ω) after modification with MB (GE/MB, curve c, Rct = 20,720 Ω), and the conductivity of the modified electrode decreased. It proves the successful modification of the MB probe on the GE through an Au–S covalent bond. After MCH was immobilized on the electrode of GE/MB, the resistance had a modest increase (curve d, Rct = 23,710 Ω). This manifested that MCH blocked the unbound sites of the electrode and reduced the electron transfer ability. However, the presence of the target initiated the HCR/CRISPR approach, leading to the cleavage of the MB probe by the activation of the Cas12a system. As a result, the resistance significantly decreased after the degradation of the MB probe on the GE (curve b, Rct = 5740 Ω). Meanwhile, CV measurements were consistent with the result of EIS measurement. The results of CV and EIS showed that this technique had a great potential to monitor the electrode surface in each step of the biosensor fabrication.
EIS (A) and CV (B) responses of the biosensor obtained by the stepwise manufacturing process for (a) bare GE, (b) MB/MCH/target, (c) MB-modified electrode, and (d) MB/MCH. Inset of A: Randles equivalent circuit for fitting impedance data. Rct, CPE, Zw, and Rs stand for the charge transfer resistance, the constant phase element, the Warburg impedance, and the ohmic resistance of the electrolyte, respectively. The EIS and CV were measured using the buffer containing 5 mM [Fe(CN)6] 3−/4− and 0.1 M KCl. The frequency range of EIS measurement was 0.1–100 kHz, and the amplitude was 5 mV. The potential range of CV measurement was – 0.2 to 0.6 V, and the scan rate was 50 mV/s
Feasibility of the electrochemical biosensor
The DPV signal was measured to verify the feasibility of the HCR-CRISPR/Cas12a approach to detect S. typhimurium. The DPV current signal was used to estimate the current signal because the HCR-CRISPR/Cas12a approach was developed based on the Cas12a system. After assembling the HCR product in the presence of the target S. typhimurium, the CRISPR-Cas12a system was directly activated, causing the degradation of the MB probe on the electrode and a significant decrease in the current signal (Fig. 3, curve d). When any element was missing (such as crRNA, target, and Cas12a), the trans-cleavage activity of Cas12a was repressive (Fig. 3 curves a, b, and c). As a result, the MB probe on the GE was preserved, with no significant change in the DPV current signal.
Optimization of the experimental conditions
In the HCR-CRISPR, the efficient trans-cleavage activity of the CRISPR-Cas12a system played an important role and affected the sensitivity in the whole experiment [24]. Hence, we first optimized the experimental conditions, such as the concentration of Cas12a and the amount of crRNA. As shown in Fig. S2, when the concentration of Cas12a was 0.3 μM and the concentration of crRNA was 1 μM, a higher enzyme digestion reaction rate was obtained (Fig. S2A and S2B). Therefore, 0.3 μM Cas12a and 1 μM crRNA were chosen as the best conditions for the CRISPR-Cas12a system. In addition, the digestion time of Lba Cas12a was monitored using the current signal of DPV. As shown in Fig. S2C, the current signal increased as the digestion time of Lba Cas12a increased from 5 to 60 min and then became stable. Therefore, 60 min was chosen as the best digestion time of Lba Cas12a for subsequent experiments.
We also optimized some other important reaction conditions to achieve a better biosensor performance in terms of the concentration of aptamer, incubation time of HCR, and amount of DBs (Fig. S3). As an important component of the recognition target, the content of the aptamer was very important for the whole experiment. A high concentration of aptamer caused steric hindrance, while a low concentration affected sensitivity. As shown in Fig. S3A, the peak current increased from 2 to 4 μM and then started to decrease. Therefore, the concentration of aptamer of 4 μM was chosen as the optimal concentration for the subsequent experiments. Then, the incubation time of HCR was examined from 5 to 75 min. As shown in Fig. S3D, the current signal value gradually increased along with the increasing HCR reaction time from 5 to 30 min and then it reaches the maximum in 60 min. Hence, the 60-min HCR incubation time was chosen to carry out the experiment. Afterward, we optimized the amount of DBs. An optimal DPV signal was monitored when the DBs was 5 μL (Fig. S3B). As a result, 5 μL of DBs was applied throughout subsequent experiments. Finally, the effect of incubation temperature as a crucial factor influencing the aptamer activity on the experiments was investigated; the results are shown in Fig. S3C. The reaction temperature increased in the range of 15–37 °C, reached a maximum at 37 °C, and then decreased with the increasing incubation temperature. This result suggested that high temperatures affected the binding efficiency of aptamer. Thus, the incubation temperature of 37 °C was used in this experiment.
Sensitivity
The sensitivity of the biosensor was studied by measuring the DPV response of a ten-fold serial dilution of S. typhimurium solution under the best-optimized conditions. As shown in Fig. 4, a gradual decrease in the DPV response was achieved by increasing the target bacteria concentration in the range of 10–108 CFU/mL. The target bacteria of S. typhimurium could be quantified in the concentration range of 104 − 108 CFU/mL. The correlation equation was Y = 0.1027*X + 0.03242 with an R2 of 0.947, where Y is the DPV current signal and X is the amount of S. typhimurium. A detection limit of 20 CFU/mL was calculated based on the 3θ/slope (θ is three times the standard deviation). A comparison with other reported CRISPR and electrochemical methods for bacterial detection is shown in Table 1. The results indicated that the limit of detection (LOD) was comparable or better than that of most other assays.
Specificity
To investigate specificity, we evaluated this assay for different foodborne pathogenic bacteria; the result is shown as a DPV response in Fig. 5. Other nontarget bacteria, including a negative control, such as E. coli, L. monocytogenes, S. aureus, and V. parahaemolyticus, had a low current response, proving the distinction specificity of this method.
Actual sample analysis
According to the national standards protocol of China (GB 4789) [29], the culture of S. typhimurium was diluted to the densities of 104, 105, and 106 CFU/mL and then added to the tenfold diluted milk samples (25 mL of milk was mixed with 225 mL of PBS buffer) for recovery measurement. As shown in Table S3, the recoveries in milk samples were 97.5–114.7%, with the relative standard deviations from 1.7 to 16.8%. The results demonstrated that this HCR-CRISPR approach possessed a broad prospect in the clinical diagnostics of real samples.
Conclusions
In this study, a rapid and sensitive electrochemical biosensor based on Cas12a-crRNA, the aptamer of pathogenic bacteria, and HCR was successfully constructed for detecting pathogenic bacteria. The results showed that the method achieved high specificity and extremely high sensitivity among different bacterial strains. The LOD was down to 20 CFU/mL. By changing the aptamer, the approach could be used on other intended bacteria. The established method could be applied to the on-site diagnosis of pathogenic bacteria using specific aptamers and the introduction of isothermal amplification HCR approach and Cas12a-crRNA [30].
References
Ansari N, Yazdian-Robati R, Shahdordizadeh M, Wang Z, Ghazvini K. Aptasensors for quantitative detection of Salmonella typhimurium. Anal Biochem. 2017;533:18–25.
Eriksson E, Aspan A. Comparison of culture, ELISA and PCR techniques for Salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet Res. 2007;3:21.
Kim JS, Lee GG, Park JS, Jung YH, Kwak HS, Kim SB, et al. A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J Food Prot. 2007;70(7):1656–62.
Cai QQ, Wang R, Qiao ZH, Yang WG. Single-digit Salmonella detection with the naked eye using bio-barcode immunoassay coupled with recombinase polymerase amplification and a CRISPR-Cas12a system. Analyst. 2021;146(17):5271–9.
Zhang YX, Wu YP, Wu YF, Chang YY, Liu M. CRISPR-Cas systems: from gene scissors to programmable biosensors. Trac-Trend Anal Chem. 2021;137.
Sheng A, Wang P, Yang J, Tang L, Chen F, Zhang J. MXene Coupled with CRISPR-Cas12a for analysis of endotoxin and bacteria. Anal Chem. 2021;93(10):4676–81.
Jiang Y, Hu M, Liu AA, Lin Y, Liu L, Yu B, et al. Detection of SARS-CoV-2 by CRISPR/Cas12a-enhanced colorimetry. ACS Sens. 2021;6(3):1086–93.
Park JS, Hsieh K, Chen LB, Kaushik A, Trick AY, Wang TH. Digital CRISPR/Cas-assisted assay for rapid and sensitive detection of SARS-CoV-2. Adv Sci. 2021;8(5).
Park BJ, Park MS, Lee JM, Song YJ. Specific detection of influenza A and B viruses by CRISPR-Cas12a-based assay. Biosensors (Basel). 2021;11(3).
Lu S, Li F, Chen Q, Wu J, Duan J, Lei X, et al. Rapid detection of African swine fever virus using Cas12a-based portable paper diagnostics. Cell Discov. 2020;6(1):18.
Park JS, Hsieh K, Chen L, Kaushik A, Trick AY, Wang TH. Digital CRISPR/Cas-assisted assay for rapid and sensitive detection of SARS-CoV-2. Adv Sci (Weinh). 2021;8(5):2003564.
Pang B, Xu J, Liu Y, Peng H, Feng W, Cao Y, et al. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal Chem. 2020;92(24):16204–12.
Xing S, Lu Z, Huang Q, Li H, Wang Y, Lai Y, et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics. 2020;10(22):10262–73.
Tian B, Minero GAS, Fock J, Dufva M, Hansen MF. CRISPR-Cas12a based internal negative control for nonspecific products of exponential rolling circle amplification. Nucleic Acids Res. 2020;48(5):e30.
Dai Y, Somoza RA, Wang L, Welter JF, Li Y, Caplan AI, et al. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal electrochemical biosensor. Angew Chem Int Ed Engl. 2019;58(48):17399–405.
Peng S, Tan Z, Chen S, Lei C, Nie Z. Integrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA. Chem Sci. 2020;11(28):7362–8.
Li F, Ye Q, Chen M, Zhou B, Zhang J, Pang R, et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens Bioelectron. 2021;179:113073.
Bi S, Yue S, Zhang S. Hybridization chain reaction: a versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem Soc Rev. 2017;46(14):4281–98.
Kachwala MJ, Smith CW, Nandu N, Yigit MV. Reprogrammable gel electrophoresis detection assay using CRISPR-Cas12a and hybridization chain reaction. Anal Chem. 2021;93(4):1934–8.
Li Y, Li SY, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37(7):730–43.
Wu H, Chen Y, Shi Y, Wang L, Zhang M, Wu J, et al. Carrying out pseudo dual nucleic acid detection from sample to visual result in a polypropylene bag with CRISPR/Cas12a. Biosens Bioelectron. 2021;178:113001.
Wei H, Bu S, Zhang W, Ma L, Liu X, Wang Z, et al. An electrochemical biosensor for the detection of pathogenic bacteria based on dual signal amplification of Cu3(PO4)2-mediated click chemistry and DNAzymes. Analyst. 2021;146(15):4841–7.
Zhang W, Bu S, Bai H, Ma C, Ma L, Wei H, et al. A sensitive biosensor for determination of pathogenic bacteria using aldehyde dehydrogenase signaling system. Anal Bioanal Chem. 2020;412(28):7955–62.
Ma L, Peng L, Yin LJ, Liu GZ, Man SL. CRISPR-Cas12a-powered dual-mode biosensor for ultrasensitive and cross-validating detection of pathogenic bacteria. Acs Sensors. 2021;6(8):2920–7.
Shen JJ, Zhou XM, Shan YY, Yue HH, Huang R, Hu JM, et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nature Communications. 2020;11(1).
Sun J, Warden AR, Huang J, Wang W, Ding X. Colorimetric and electrochemical detection of Escherichia coli and antibiotic resistance based on a p-benzoquinone-mediated bioassay. Anal Chem. 2019;91(12):7524–30.
Feng Y, Zhou D, Gao L, He F. Electrochemical biosensor for rapid detection of bacteria based on facile synthesis of silver wire across electrodes. Biosens Bioelectron. 2020;168:112527.
Nemr CR, Smith SJ, Liu W, Mepham AH, Mohamadi RM, Labib M, et al. Nanoparticle-mediated capture and electrochemical detection of methicillin-resistant Staphylococcus aureus. Anal Chem. 2019;91(4):2847–53.
Zhan LZ, Song DF, Gu Q, Yan TT, Ma CC. Reverse transcription - loop-mediated isothermal amplification assay for the rapid detection of pathogenic Listeria monocytogenes in meat products. Can J Microbiol. 2019;65(12):913–21.
Wang Y, Ke Y, Liu W, Sun Y, Ding X. A one-pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sens. 2020;5(5):1427–35.
Funding
This work was financially supported by the Science and Technology Development Project of Jilin Province (20150101105JC).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiu Liu and Shengjun Bu contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, X., Bu, S., Feng, J. et al. Electrochemical biosensor for detecting pathogenic bacteria based on a hybridization chain reaction and CRISPR-Cas12a. Anal Bioanal Chem 414, 1073–1080 (2022). https://doi.org/10.1007/s00216-021-03733-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03733-6