Abstract
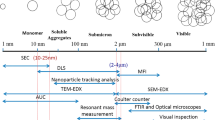
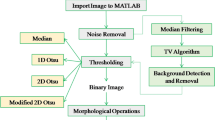
Aggregation of therapeutic proteins is a major concern as aggregates lower the yield and can impact the efficacy of the drug as well as the patient’s safety. It can occur in all production stages; thus, it is essential to perform a detailed analysis for protein aggregates. Several methods such as size exclusion high-performance liquid chromatography (SE-HPLC), light scattering, turbidity, light obscuration, and microscopy-based approaches are used to analyze aggregates. None of these methods allows determination of all types of higher molecular weight (HMW) species due to a limited size range. Furthermore, quantification and specification of different HMW species are often not possible. Moreover, automation is a perspective challenge coming up with automated robotic laboratory systems. Hence, there is a need for a fast, high-throughput-compatible method, which can detect a broad size range and enable quantification and classification. We describe a novel approach for the detection of aggregates in the size range 1 to 1000 μm combining fluorescent dyes for protein aggregate labelling and automated fluorescence microscope imaging (aFMI). After appropriate selection of the dye and method optimization, our method enabled us to detect various types of HMW species of monoclonal antibodies (mAbs). Using 10 μmol L−1 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonate (Bis-ANS) in combination with aFMI allowed the analysis of mAb aggregates induced by different stresses occurring during downstream processing, storage, and administration. Validation of our results was performed by SE-HPLC, UV-Vis spectroscopy, and dynamic light scattering. With this new approach, we could not only reliably detect different HMW species but also quantify and classify them in an automated approach. Our method achieves high-throughput requirements and the selection of various fluorescent dyes enables a broad range of applications.



Similar content being viewed by others
References
Leavy O. Therapeutic antibodies: past, present and future. Nat Rev Immunol. 2010;10(5):297. doi:10.1038/nri2763.
Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science (New York, NY). 2013;341(6151):1192–8. doi:10.1126/science.1241145.
Kuystermans D, Al-Rubeai M. Biopharmaceutical products from animal cell culture. In: Al-Rubeai M, editor. Animal cell culture. Cham: Springer International Publishing; 2015. p. 717–57. doi:10.1007/978-3-319-10320-4_23.
Vázquez-Rey M, Da L. Aggregates in monoclonal antibody manufacturing processes. Biotechnol Bioeng. 2011;108(7):1494–508. doi:10.1002/bit.23155.
Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. J Biol Chem. 2011;286(28):25118–33. doi:10.1074/jbc.M110.160457.
Roberts CJ. Protein aggregation and its impact on product quality. Curr Opin Biotechnol. 2014;30:211–7. doi:10.1016/j.copbio.2014.08.001.
Paul AJ, Schwab K, Hesse F. Direct analysis of mAb aggregates in mammalian cell culture supernatant. BMC Biotechnol. 2014;14(1):99. doi:10.1186/s12896-014-0099-3.
Cromwell MEM, Hilario E, Jacobson F. Protein aggregation and bioprocessing. AAPS J. 2006;8(3):E572–9. doi:10.1208/aapsj080366.
den Engelsman J, Garidel P, Smulders R, Koll H, Smith B, Bassarab S, Seidl A, Hainzl O, Jiskoot W. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm Res. 2011;28(4):920–33. doi:10.1007/s11095-010-0297-1.
Das TK. Protein particulate detection issues in biotherapeutics development—current status. AAPS PharmSciTech. 2012;13(2):732–46. doi:10.1208/s12249-012-9793-4.
Zolls S, Tantipolphan R, Wiggenhorn M, Winter G, Jiskoot W, Friess W, Hawe A. Particles in therapeutic protein formulations, part 1: overview of analytical methods. J Pharm Sci. 2012;101(3):914–35. doi:10.1002/jps.23001.
Ripple DC, Montgomery CB, Hu Z. An interlaboratory comparison of sizing and counting of subvisible particles mimicking protein aggregates. J Pharm Sci. 2015;104(2):666–77. doi:10.1002/jps.24287.
Weinbuch D, Jiskoot W, Hawe A. Light obscuration measurements of highly viscous solutions: sample pressurization overcomes underestimation of subvisible particle counts. AAPS J. 2014;16(5):1128–31. doi:10.1208/s12248-014-9629-0.
Demeule B, Messick S, Shire SJ, Liu J. Characterization of particles in protein solutions: reaching the limits of current technologies. AAPS J. 2010;12(4):708–15. doi:10.1208/s12248-010-9233-x.
Barnard JG, Singh S, Randolph TW, Carpenter JF. Subvisible particle counting provides a sensitive method of detecting and quantifying aggregation of monoclonal antibody caused by freeze-thawing: insights into the roles of particles in the protein aggregation pathway. J Pharm Sci. 2011;100(2):492–503. doi:10.1002/jps.22305.
Huang C-T, Sharma D, Oma P, Krishnamurthy R. Quantitation of protein particles in parenteral solutions using micro-flow imaging. J Pharm Sci. 2009;98(9):3058–71. doi:10.1002/jps.21575.
Lawler DM. Turbidimetry and nephelometry. In: Worsfold PJT, Townshend A, Poole, CF, editors. Encyclopedia of analytical sciences. Elsevier; p 343–351. doi:10.1016/B0-12-369397-7/00718-4.
Arakawa T, Philo JS, Ejima D, Tsumoto K, Arisaka F. Aggregation analysis of therapeutic proteins, part 2: analytical ultracentrifugation and dynamic light scattering. Bioprocess Int. 2007;5:36–47.
Demeule B, Gurny R, Arvinte T. Detection and characterization of protein aggregates by fluorescence microscopy. Int J Pharm. 2007;329(1–2):37–45. doi:10.1016/j.ijpharm.2006.08.024.
Paul AJ, Schwab K, Prokoph N, Haas E, Handrick R, Hesse F. Fluorescence dye-based detection of mAb aggregates in CHO culture supernatants. Anal Bioanal Chem. 2015; doi:10.1007/s00216-015-8672-8.
Rohm M, Handl A, Konig M, Mavoungou C, Handrick R, Schindowski K. Data of rational process optimization for the production of a full IgG and its Fab fragment from hybridoma cells. Data Brief. 2016;8:426–35. doi:10.1016/j.dib.2016.05.067.
Katayama DS, Nayar R, Chou DK, Campos J, Cooper J, Vander Velde DG, Villarete L, Liu CP, Cornell Manning M. Solution behavior of a novel type 1 interferon, interferon-tau. J Pharm Sci. 2005;94(12):2703–15. doi:10.1002/jps.20461.
Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies—application of platform approaches. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848(1):28–39. doi:10.1016/j.jchromb.2006.09.026.
Joshi V, Shivach T, Kumar V, Yadav N, Rathore A. Avoiding antibody aggregation during processing: establishing hold times. Biotechnol J. 2014;9(9):1195–205. doi:10.1002/biot.201400052.
Yoshimura Y, Lin Y, Yagi H, Lee YH, Kitayama H, Sakurai K, So M, Ogi H, Naiki H, Goto Y. Distinguishing crystal-like amyloid fibrils and glass-like amorphous aggregates from their kinetics of formation. Proc Natl Acad Sci U S A. 2012;109(36):14446–51. doi:10.1073/pnas.1208228109.
Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1(5):639–48.
Bickel F, Herold EM, Signes A, Romeijn S, Jiskoot W, Kiefer H. Reversible NaCl-induced aggregation of a monoclonal antibody at low pH: characterization of aggregates and factors affecting aggregation. Eur J Pharm Biopharm. 2016;107:310–20. doi:10.1016/j.ejpb.2016.07.020.
Hawe A, Kasper JC, Friess W, Jiskoot W. Structural properties of monoclonal antibody aggregates induced by freeze-thawing and thermal stress. Eur J Pharm Sci. 2009;38(2):79–87. doi:10.1016/j.ejps.2009.06.001.
Kueltzo LA, Wang WEI, Randolph TW, Carpenter JF. Effects of solution conditions, processing parameters, and container materials on aggregation of a monoclonal antibody during freeze-thawing. J Pharm Sci. 2008;97(5):1801–12. doi:10.1002/jps.
Miller MA, Khan TA, Kaczorowski KJ, Wilson BK, Dinin AK, Borwankar AU, Rodrigues MA, Truskett TM, Johnston KP, Maynard JA. Antibody nanoparticle dispersions formed with mixtures of crowding molecules retain activity and in vivo bioavailability. J Pharm Sci. 2012;101(10):3763–78. doi:10.1002/jps.23256.
Respaud R, Marchand D, Parent C, Pelat T, Thullier P, Tournamille JF, Viaud-Massuard MC, Diot P, Si-Tahar M, Vecellio L, Heuze-Vourc'h N. Effect of formulation on the stability and aerosol performance of a nebulized antibody. MAbs. 2014;6(5):1347–55. doi:10.4161/mabs.29938.
Siekmeier R, Scheuch G. Systemic treatment by inhalation of macromolecules—principles, problems, and examples. J Physiol Pharmacol. 2008;59(Suppl. 6):53–79.
Maa YF, Hsu CC. Protein denaturation by combined effect of shear and air-liquid interface. Biotechnol Bioeng. 1997;54(6):503–12. doi:10.1002/(SICI)1097-0290(19970620)54:6<503::AID-BIT1>3.0.CO;2-N.
Yano YF, Uruga T, Tanida H, Toyokawa H, Terada Y, Takagaki M, Yamada H. Driving force behind adsorption-induced protein unfolding: a time-resolved X-ray reflectivity study on lysozyme adsorbed at an air/water interface. Langmuir. 2009;25(1):32–5. doi:10.1021/la803235x.
Vermeer AW, Norde W. The thermal stability of immunoglobulin: unfolding and aggregation of a multi-domain protein. Biophys J. 2000;78(1):394–404. doi:10.1016/S0006-3495(00)76602-1.
Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi:10.1007/s11095-010-0073-2.
Zolls S, Weinbuch D, Wiggenhorn M, Winter G, Friess W, Jiskoot W, Hawe A. Flow imaging microscopy for protein particle analysis—a comparative evaluation of four different analytical instruments. AAPS J. 2013;15(4):1200–11. doi:10.1208/s12248-013-9522-2.
Hawe A, Schaubhut F, Geidobler R, Wiggenhorn M, Friess W, Rast M, de Muynck C, Winter G. Pharmaceutical feasibility of sub-visible particle analysis in parenterals with reduced volume light obscuration methods. Eur J Pharm Biopharm. 2013;85(3 Pt B):1084–7. doi:10.1016/j.ejpb.2013.02.004.
Hertel S, Pohl T, Friess W, Winter G. That’s cool!—nebulization of thermolabile proteins with a cooled vibrating mesh nebulizer. Eur J Pharm Biopharm. 2014;87(2):357–65. doi:10.1016/j.ejpb.2014.03.001.
Acknowledgments
This research was supported by the German Federal Ministry of Education and Research (Grant No. 0315342A) and by the Cooperative Research Training Group Pharmaceutical Biotechnology stated by the Postgraduate Scholarships Act of the Ministry for Science, Research, and Arts of the federal state government of Baden-Württemberg. Further funding was obtained by the German Research Foundation (DFG) through the International Graduate School in Molecular Medicine at Ulm University. In addition, the authors are grateful to Rentschler Biotechnologie GmbH (Laupheim, Germany) for providing mAb1 and to Boehringer Ingelheim Pharma GmbH & Co. KG (Ingelheim, Germany) for offering mAb2 and mAb3. The mAb4 was kindly donated by Kenneth Siddle (Cambridge University, England).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
ESM 1
(PDF 1897 kb)
Rights and permissions
About this article
Cite this article
Paul, A.J., Bickel, F., Röhm, M. et al. High-throughput analysis of sub-visible mAb aggregate particles using automated fluorescence microscopy imaging. Anal Bioanal Chem 409, 4149–4156 (2017). https://doi.org/10.1007/s00216-017-0362-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0362-2