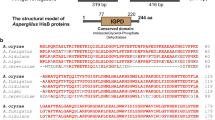
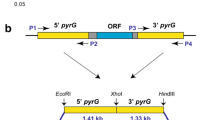
Abstract
The filamentous fungus Aspergillus niger is widely exploited as an industrial workhorse for producing enzymes and organic acids. So far, different genetic tools, including CRISPR/Cas9 genome editing strategies, have been developed for the engineering of A. niger. However, these tools usually require a suitable method for gene transfer into the fungal genome, like protoplast-mediated transformation (PMT) or Agrobacterium tumefaciens-mediated transformation (ATMT). Compared to PMT, ATMT is considered more advantageous because fungal spores can be used directly for genetic transformation instead of protoplasts. Although ATMT has been applied in many filamentous fungi, it remains less effective in A. niger. In the present study, we deleted the hisB gene and established an ATMT system for A. niger based on the histidine auxotrophic mechanism. Our results revealed that the ATMT system could achieve 300 transformants per 107 fungal spores under optimal transformation conditions. The ATMT efficiency in this work is 5 − 60 times higher than those of the previous ATMT studies in A. niger. The ATMT system was successfully applied to express the DsRed fluorescent protein-encoding gene from the Discosoma coral in A. niger. Furthermore, we showed that the ATMT system was efficient for gene targeting in A. niger. The deletion efficiency of the laeA regulatory gene using hisB as a selectable marker could reach 68 − 85% in A. niger strains. The ATMT system constructed in our work represents a promising genetic tool for heterologous expression and gene targeting in the industrially important fungus A. niger.
Graphical abstract





Similar content being viewed by others
Data availability
All data for this study are included in the manuscript and its supplementary material file.
References
Basenko EY, Pulman JA, Shanmugasundram A, Harb OS, Crouch K, Starns D, Warrenfeltz S, Aurrecoechea C Jr, Stoeckert CJ, Kissinger JC, Roos DS, Hertz-Fowler C (2018) FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J. Fungi 4:39
Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, Slakhorst SM (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet 14:437–443
Busch S, Hoffmann B, Valerius O, Starke K, Düvel K, Braus GH (2001) Regulation of the Aspergillus nidulans hisB gene by histidine starvation. Curr Genet 38:314–322
Cairns TC, Nai C, Meyer V (2018) How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol Biotechnol 5:13
de Groot MJ, Bundock P, Hooykaas PJ, Beijersbergen AG (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16:839–842
Dietl AM, Amich J, Leal S, Beckmann N, Binder U, Beilhack A, Pearlman E, Haas H (2016) Histidine biosynthesis plays a crucial role in metal homeostasis and virulence of Aspergillus fumigatus. Virulence 7:465–476
Fieldler MRM, Gensheimer T, Kubisch C, Meyer V (2017) HisB as novel selection marker for gene targeting approaches in Aspergillus niger. BMC Microbiol 17:57
Frandsen RJ (2011) A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J Microbiol Methods 87:247–262
Idnurm A, Bailey AM, Cairns TC, Elliott CE, Foster GD, Ianiri G, Jeon J (2017) A silver bullet in a golden age of functional genomics: the impact of Agrobacterium-mediated transformation of fungi. Fungal Biol Biotechnol 4:6
Jain S, Keller N (2013) Insights to fungal biology through LaeA sleuthing. Fungal Biol Rev 27:51–59
Jeong JY, Yim HS, Ryu JY, Lee HS, Lee JH, Seen DS, Kang SG (2012) One-step Sequence- and Ligation-Independent Cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol 78:5440–5443
Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Müller WH, Dijksterhuis J, Wösten HA (2013) Development in Aspergillus. Stud Mycol 74:1–29
Li M, Zhou L, Liu M, Huang Y, Sun X, Lu F (2013) Construction of an engineering strain producing high yields of α-transglucosidase via Agrobacterium tumefaciens-mediated transformation of Aspergillus niger. Biosci Biotechnol Biochem 77:1860–1866
Li D, Tang Y, Lin J, Cai W (2017) Methods for genetic transformation of filamentous fungi. Microb Cell Fact 16:168
Li C, Zhou J, Du G, Chen J, Takahashi S, Liu S (2020) Developing Aspergillus niger as a cell factory for food enzyme production. Biotechnol Adv 44:107630
Liu D, Liu Q, Guo W, Liu Y, Wu M, Zhang Y, Li J, Sun W, Wang X, He Q, Tian C (2022) Development of genetic tools in glucoamylase-hyperproducing industrial Aspergillus niger strains. Biology 11:1396
Meijer M, Houbraken JA, Dalhuijsen S, Samson RA, de Vries RP (2011) Growth and hydrolase profiles can be used as characteristics to distinguish Aspergillus niger and other black Aspergilli. Stud Mycol 69:19–30
Meyer V, Wu B, Ram AF (2011) Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol Lett 33:469–476
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005a) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17
Michielse CB, Arentshorst M, Ram AF, van den Hondel CA (2005b) Agrobacterium-mediated transformation leads to improved gene replacement efficiency in Aspergillus awamori. Fungal Genet Biol 42:9–19
Mora-Lugo R, Zimmermann J, Rizk AM, Fernandez-Lahore M (2014) Development of a transformation system for Aspergillus sojae based on the Agrobacterium tumefaciens-mediated approach. BMC Microbiol 14:247
Nguyen KT, Ho QN, Pham TH, Phan TN, Tran VT (2016) The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium-mediated transformation of Aspergillus oryzae. World J Microbiol Biotechnol 32:204
Nguyen TK, Ho NQ, Do TBXL, Mai LTD, Pham DN, Tran HTT, Le DH, Nguyen HQ, Tran VT (2017) A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J Microbiol Biotechnol 33:107
Niu J, Arentshorst M, Nair PD, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AF (2015) Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 Bethesda 6:193–204
Pronk JT (2002) Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 68:2095–2100
Son YE, Park HS (2021) Genetic manipulation and transformation methods for Aspergillus spp. Mycobiology 49:95–104
Sugui JA, Chang YC, Kwon-Chung KJ (2005) Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Appl Environ Microbiol 71:1798–1802
Thai HD, Nguyen BPT, Nguyen VM, Nguyen QH, Tran VT (2021) Development of a new Agrobacterium-mediated transformation system based on a dual auxotrophic approach in the filamentous fungus Aspergillus oryzae. World J Microbiol Biotechnol 37:92
Ullah M, Xia L, Xie S, Sun S (2020) CRISPR/Cas9-based genome engineering: A new breakthrough in the genetic manipulation of filamentous fungi. Biotechnol Appl Biochem 67:835–851
Weld RJ, Plummer KM, Carpenter MA, Ridgway HJ (2006) Approaches to functional genomics in filamentous fungi. Cell Res 16:31–44
Weyda I, Yang L, Vang J, Ahring BK, Lübeck M, Lübeck PS (2017) A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J Microbiol Methods 135:26–34
Yang L, Lübeck M, Lübeck PS (2017) Aspergillus as a versatile cell factory for organic acid production. Fungal Biol Rev 31:33–49
Funding
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.04-2018.36. Hanh-Dung Thai was funded by Vingroup JSC and supported by the Master, PhD Scholarship Programme of Vingroup Innovation Foundation (VINIF), Institute of Big Data, code VINIF.2021.TS.076.
Author information
Authors and Affiliations
Contributions
VTT and HDT designed the study. HDT, LTBXD, XTN, TXV, and HTTT performed experiments. HDT, HQN, and VTT conducted the data analysis. VTT and HDT wrote the manuscript. VTT and HQN edited the final manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thai, HD., Do, L.T.B.X., Nguyen, X.T. et al. A newly constructed Agrobacterium-mediated transformation system based on the hisB auxotrophic marker for genetic manipulation in Aspergillus niger. Arch Microbiol 205, 183 (2023). https://doi.org/10.1007/s00203-023-03530-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03530-y