Abstract
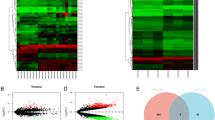
We have previously shown that circRNAs in host cells are involved in the process of Chlamydia trachomatis infection. In this study we aimed to identify significantly altered circRNAs/lncRNAs/mRNAs in Chlamydia muridarum infected cells and investigate their biological functions in the interaction between Chlamydia muridarum and host cells. For this purpose, circRNA, lncRNA and mRNA expression profiles were screened and identified in HeLa cells with or without Chlamydia muridarum infection by microarray. Bioinformatics analyses including Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis were then carried out and the circRNA-miRNA ceRNA network was constructed. The differentially expressed circRNAs and lncRNAs were selected for validation by RT-qPCR. The results shown that a total of 834 circRNAs, 2149 lncRNAs and 1283 mRNAs were found to be differentially expressed. Enrichment analysis of GO and KEGG showed that the dysregulated genes involved nuclear-transcribed mRNA catabolic process, protein binding, RNA catabolic process and translation, the MAPK signaling pathway, apoptosis, Toll-like receptor signaling pathway, cAMP signaling pathway and Notch signaling pathway may play important roles in Chlamydia infection. Our study provides a systematic outlook on the potential function of non-coding RNAs in the molecular basis of Chlamydia infection.





Similar content being viewed by others
References
Arkatkar T, Gupta R, Li W, Yu JJ, Wali S, Neal GM, Chambers JP, Christenson LK, Arulanandam BP (2015) Murine MicroRNA-214 regulates intracellular adhesion molecule (ICAM1) gene expression in genital Chlamydia muridarum infection. Immunology 145:534–542
Benyeogor I, Simoneaux T, Wu Y, Lundy S, George Z, Ryans K, McKeithen D, Pais R, Ellerson D, Lorenz WW et al (2019) A unique insight into the MiRNA profile during genital chlamydial infection. BMC Genomics 20:143
Centers for Disease Control and Prevention (2019) Sexually transmitted disease surveillance 2018[R]. Atlanta: U.S. Department of Health and Human Services. pp 4
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388
Dong F, Pirbhai M, Xiao Y, Zhong Y, Wu Y, Zhong G (2005) Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect Immun 73:1861–1864
Fan T, Lu H, Hu H, Shi L, McClarty GA, Nance DM, Greenberg AH, Zhong G (1998) Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J Exp Med 187:487–496
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hu C, Wu H, Sun Y, Kong J, Shao L, Chen X, Liu Q, Liu Y (2020) GlgA plays an important role in the induction of hydrosalpinx by Chlamydia muridarum. Pathog Dis 78:ftaa027
Huang Y (2018) The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med 22:5768–5775
Huang Y, Sun Y, Qin T, Liu Y (2019) The Structural integrity of plasmid-encoded Pgp3 Is essential for induction of hydrosalpinx by Chlamydia muridarum. Front Cell Infect Microbiol 9:13
Kun D, Xiang-Lin C, Ming Z, Qi L (2013) Chlamydia inhibit host cell apoptosis by inducing Bag-1 via the MAPK/ERK survival pathway. Apoptosis 18:1083–1092
Li M, Meng F, Lu Q (2020) Expression profile screening and bioinformatics analysis of circRNA, LncRNA, and mRNA in acute myeloid leukemia drug-resistant cells. Turk J Haematol 37:104–110
Liu Y, Huang Y, Yang Z, Sun Y, Gong S, Hou S, Chen C, Li Z, Liu Q, Wu Y, Baseman J, Zhong G (2014) Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect Immun 82:5327–5335
Liu Y, Hu C, Sun Y, Wu H, Chen X, Liu Q (2019) Identification of differentially expressed circular RNAs in HeLa cells infected with Chlamydia trachomatis. Pathog Dis 77:ftz062
Lundy SR, Abney K, Ellerson D, Igietseme JU, Carroll D, Eko FO, Omosun YO (2021) MiR-378b modulates chlamydia-induced upper genital tract pathology. Pathogens 10:566
Mukherjee S, Akbar I, Kumari B, Vrati S, Basu A, Banerjee A (2019) Japanese encephalitis virus-induced let-7a/b interacted with the NOTCH-TLR7 pathway in microglia and facilitated neuronal death via caspase activation. J Neurochem 149:518–534
Ouyang J, Zhu X, Chen Y, Wei H, Chen Q, Chi X, Qi B, Zhang L, Zhao Y, Gao GF et al (2014) NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16:616–626
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:141–148
Rajalingam K, Sharma M, Paland N, Hurwitz R, Thieck O, Oswald M, Machuy N, Rudel T (2006) IAP-IAP complexes required for apoptosis resistance of C. trachomatis-infected cells. PLoS Pathog 2:e114
Robinson EK, Covarrubias S, Carpenter S (2020) The how and why of lncRNA function: an innate immune perspective. Biochim Biophys Acta Gene Regul Mech 1863:194419
Sun Y, Kong J, Ma J, Qi M, Zhang Y, Han L, Liu Q, Liu Y (2018) Chlamydial genomic MinD protein does not regulate plasmid-dependent genes like Pgp5. Acta Biochim Pol 65:425–429
Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A (2006) Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34:e63
Wen Y, Chen H, Luo F, Zhao L, Shu M, Su S, Zhao Y, Huang Q, Li Z (2020) Chlamydia trachomatis plasmid protein pORF5 up-regulates ZFAS1 to promote host cell survival via MAPK/p38 pathway. Front Microbiol 11:593295
Xiao Y, Zhong Y, Greene W, Dong F, Zhong G (2004) Chlamydia trachomatis infection inhibits both Bax and Bak activation induced by staurosporine. Infect Immun 72:5470
Yan L, Yanbing Z, Xiao X, Mona S, Ke L, Jianchao W, Donghua S, Beibei L, Guangzhi T, Michal AO et al (2020) Notch signaling contributes to the expression of inflammatory cytokines induced by highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infection in porcine alveolar macrophages. Dev Comp Immunol 108:103690
Yeruva L, Pouncey DL, Eledge MR, Bhattacharya S, Luo C, Weatherford EW, Ojcius DM, Rank RG (2017) MicroRNAs modulate pathogenesis resulting from chlamydial infection in mice. Infect Immun 85:e00768-16
Yue X, Gong X, Li J, Zhang J (2020) Epidemiologic features of genital Chlamydia trachomatis infection at national sexually transmitted disease surveillance sites in China, 2015–2019. Chin J Dermatol 53(08):596–601
Zhang Q, Chen CY, Yedavalli VS, Jeang KT (2013) NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. Mbio 4:e512–e596
Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ (2018) CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci 22:2297–2303
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31570178, 31100138) and the Natural Science Foundation of Tianjin (Grant No. 16JCQNJC11100). In addition, we thank the researchers in the Shanghai KangChen Biotech for their technical support.
Author information
Authors and Affiliations
Contributions
YL, HW and YS conceived and designed the experiments; YL, JX and XH performed the experiments; HW and YS analyzed the data; YL and YS wrote the paper.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Xiang, J., Hu, X. et al. Expression profile screening and bioinformatics analysis of CircRNA, LncRNA, and mRNA in HeLa cells infected with Chlamydia muridarum. Arch Microbiol 204, 352 (2022). https://doi.org/10.1007/s00203-022-02941-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02941-7