Abstract
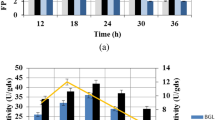
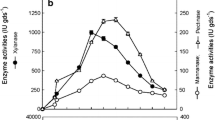
Fruit and vegetable wastes create unhygienic conditions and pose a environmental pollution. The utilization of such wastes as carbon sources for production of enzyme with microbial intervention could be an ecofriendly and profitable approach, apart from diminishing the waste load. The present investigation focused on the feasibility of using mosambi (Citrus limetta) peel as substrate for multienzyme production (pectinase, cellulase and amylase) through microbial intervention. Fifteen fungi were isolated from organic waste and screened in vitro their potential of biodegradation of mosambi peel through enzymes production. The best performing isolate was selected and identified as Trichoderma asperellum NG-125 (accession number-MW287256). Conditions viz. temperature, pH, incubation time and nutrient addition were optimized for efficient enzymes production. The maximum enzyme activity (U ml−1 min−1) of pectinase (595.7 ± 2.47), cellulase (497.3 ± 2.06) and amylase (440.9 ± 1.44) were observed at pH 5.5, incubation temperature of 30 °C after 10 days of fermentation. Moreover, macro-nutrients such as ammonium sulfate (0.1%) and potassium-di-hydrogen-ortho-phosphate (0.01%) further also enhanced the production of enzymes. The SDS-PAGE analysis of purified pectinase, cellulase and amylase using showed molecular mass of 43, 66 and 33 kDa, respectively. The enzyme retention activity (ERA) of aforesaid enzymes was also tested with four different natural fiber matrices viz., bagasse, rice husk, paddy straw and wheat straw. Among these, the maximum ERA was observed on bagasse matrix (pectinase—56.35%, cellulose—77.68% and amylase 59.54%). Enzymatic juice clarification yield obtained with test enzyme was 75.8%, as compared to 80.5% of commercial enzyme. The result indicates that T. asperellum may be exploited as multifaceted biocatalysis.









Similar content being viewed by others
References
Abdulaal WH (2018) Purification and characterization of α-amylase from Trichoderma pseudokoningii. BMC Biochem 19:4. https://doi.org/10.1186/s12858-018-0094-8
Aravantinos-Zafiris G, Oreopoulou V, Tzia C, Thomopoulos CD (1994) Fibre fraction from orange peel residues after pectin extraction. Lebensm-Wiss u-Technol 27:468–471. https://doi.org/10.1006/FSTL.1994.1094
Arotupin DJ, Akinyosoye FA, Onifade AK (2008) Purification and characterization of pectinmethylesterase from Aspergillus repens isolated from cultivated soil. Afr J Biotechnol 7:1991–1998. https://doi.org/10.5897/AJB2008.000-5046
Azam Z, Ayaz A, Younas M, Qureshi Z, Arshad B, Zaman W, Ullah F, Nasar MQ, Bahadur S, Irfan MM, Hussain S, Saqib S (2020) Microbial synthesized cadmium oxide nanoparticles induce oxidative stress and protein leakage in bacterial cells. Microb Pathog. https://doi.org/10.1016/j.micpath.2020.104188
Babu VG, Perumal P, Muthu S, Pichai S, Sankar NK, Malairaj S (2018) Enhanced method for high spatial resolution surface imaging and analysis of fungal spores using scanning electron microscopy. Sci Rep 2(8):16278. https://doi.org/10.1038/s41598-018-34629-8
Banu AR, Devi MK, Gnanaprabhal GR, Pradeep BV, Palaniswamiy M (2010) Production and characterization of pectinase enzyme from Penicillium chrysogenum. Indian J Sci Technol 3:377–381
Bech L, Busk PK, Lange L (2014) Cell wall degrading enzymes in Trichoderma asperellum grown on wheat bran. Fungal Genom Biol 4:116. https://doi.org/10.4172/2165-8056.1000126
Bech L, Herbst FA, Grell MN, Hai Z, Lange L (2015) On-Site enzyme production by Trichoderma asperellum for the degradation of duckweed. Fungal Genom Biol 5:126. https://doi.org/10.4172/2165-8056.1000116
Dai J, Dong A, Xiong G, Liu Y, Hossain MS, Liu S, Qiu D (2020) Production of highly active extracellular amylase and cellulase from Bacillus subtilis ZIM3 and a recombinant strain with a potential application in tobacco fermentation. Front Microbiol 11:1539. https://doi.org/10.3389/fmicb.2020.01539
Elsababty Z, Ali AM, Houbraken J (2015) Cellulolytic and pectinolytic enzymes of some selected heat resistant fungi. J Microbiol Exp 2:66–69. https://doi.org/10.15406/jmen.2015.02.00042
Gautam SP, Bundela PS, Pandey AK, Khan J, Awasthi MK, Sarsaiya S (2011) Optimization for the production of cellulase enzyme from municipal solid waste residue by two novel cellulolytic fungi. Biotechnol Res Int. https://doi.org/10.4061/2011/810425
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research, 2nd edn. Wiley, NewYork
Horwitz W, Latimer GW (2005) AOAC Official method of analysis of 18th ed. Gaithersburg, Md. AOAC International
Juwon AD, Emmanuel OF (2012) Experimental investigations on the effects of carbon and nitrogen sources on concomitant amylase and polygalacturonase production by Trichoderma viride BITRS-1001 in submerged fermentation. Biotechnol Res Int. https://doi.org/10.1155/2012/904763
Kumar DP, Thangabalan B, Rao PV, Yugandha NM (2011) Production of pectinase enzyme by Aspergillus niger using Ficus religiosa leaves in solid state fermentation. Int J Pharm Technol 3:1351–1359
Laemmli UK (1970) Cleavage of structure protein during the assembly of the head of bacteriophase T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Levin L, Melignani E, Ramos AM (2010) Effect of nitrogen sources and vitamins on ligninolytic enzyme production by some white-rot fungi dye decolorization by selected culture filtrates. Bioresour Technol 101:4554–4563. https://doi.org/10.1016/j.biortech.2010.01.102
Li PJ, Xia JL, Shan Y, Nie ZY (2015) Comparative study of multienzyme production from typical agro-industrial residues and ultrasound-assisted extraction of crude enzyme in fermentation with Aspergillus japonicus PJ01. Bioprocess Biosyst Eng 38:2013–2022. https://doi.org/10.1007/s00449-015-1442-3
Liaquat F, Munis MFH, Haroon U, Arif S, Saqib S, Zaman W, Khan AR, Shi J, Che S, Liu Q (2020) Evaluation of metal tolerance of fungal strains isolated from contaminated mining soil of Nanjing, China. Biology 9:469. https://doi.org/10.3390/biology9120469
Maria R, Romero-Lopez P, Luis A, Bello-Perez JT, Aurea BN (2011) Fiber concentrate from orange (citrus sinensis) bagasse: characterization and application as bakery product ingredient. Int J Mol Sci 12:2174–2186. https://doi.org/10.3390/ijms12042174
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. https://doi.org/10.1021/ac60147a030
Mohamed SA, Al-Malki AL, Kumosani TA (2009) Characterization of a polygalacturonase from Trichoderma harzianum grown on citrus peel with application for apple juice. Aust J Basic Appl Sci 3:2770–2777
Nabi NG, Asgher M, Shah AH, Sheikh MA, Asad MJ (2003) Production of pectinase by Trichoderma harzianum in solid state fermentation of Citrus peel. Pak J Agric Sci 40:3–4
Nagy S, Shaw P, Veldhuis MK (1977) Citrus science and technology, vol 1. The AVI Publishing Company Inc, Westport, pp 74–427
Nathan VK, Rani ME, Rathinasamy G, Kannan ND (2017) Low molecular weight xylanase from Trichoderma viride VKF3 for bio-bleaching of newspaper pulp. BioResources 12:5264–5278. https://doi.org/10.15376/biores.12.3.5264-5278
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Mengistu F, Pagadala VK (2017) Isolation and screening of amylase producing thermophilic spore-forming Bacilli from starch-rich soil and characterization of their amylase activity. Afr J Microbiol Res 11(21): 851-859. https://doi.org/10.5897/AJMR2017.8543
Ranganna S (2001) Handbook of analysis and quality control for fruits and vegetable products, 7th edn. Tata McGraw Hill Book Co, New Delhi, pp 594–625
Raveendran S, Parameswaran B, Ummalyma SB, Abraham A, Mathew AK, Madhavan A, Rebello S, Pandey A (2018) Applications of microbial enzymes in food industry. Food Technol Biotechnol 56:16–30. https://doi.org/10.17113/ftb.56.01.18.5491
Ravindran R, Hassan SS, Williams GA, Jaiswal AK (2018) A review on bioconversion of agro-industrial wastes to industrially important enzymes. Bioengineering (Basel Switzerland) 5:93. https://doi.org/10.3390/bioengineering5040093
Ruiz HA, Rodríguez-Jasso RM, Rodríguez R, Contreras-Esquivel JC, Aguilar CN (2012) Pectinase production from lemon peel pomace as support and carbon source in solid-state fermentation column-tray bioreactor. Biochem Eng J 65:90–95. https://doi.org/10.1016/j.bej.2012.03.007
Sagar NA, Pareek S, Sharma S, Elhadi M, Yahia MLG (2018) Fruit and vegetable waste: bioactive compounds, their extraction, and possible utilization. Compr Rev Food Sci Food Saf 17:512–531. https://doi.org/10.1111/1541-4337.12330
Sazci A, Radford A, Erenlerr K (1986) Detection of cellulolytic fungi by using congo red as an indicator: a comparative study with the dinitro salicylic acid reagent method. J Appl Microbiol 61:559–562. https://doi.org/10.1111/j.1365-2672.1986.tb01729.x
Shariq M, Sohail M (2019) Citrus limetta peels: a promising substrate for the production of multienzyme preparation from a yeast consortium. Bioresour Bioprocess 6:43. https://doi.org/10.1186/s40643-019-0278-0
Sharma R, Oberoi HS, Dhillon GS (2017) Fruit and vegetable processing waste: renewable feed stocks for enzyme production. In: Dhillon GS, Kaur S (eds) agro-industrial wastes as feedstock for enzyme production. Academic Press, pp 23–59
Singh S, Mandal SK (2012) Optimization of processing parameters for production of pectinolytic enzymes from fermented pineapple residue of mixed Aspergillus species. Jordan J Biol Sci 5:307–314. https://doi.org/10.1016/B978-0-12-802392-1.00002-2
Sreena CP, Sebastian D (2018) Augmented cellulase production by Bacillus subtilis strain MU S1 using different statistical experimental designs. J Genet Eng Biotechnol 16:9–16. https://doi.org/10.1016/j.jgeb.2017.12.005
Sudeep KC, Upadhyaya J, Joshi DR, Lekhak B, Chaudhary DK, Pant BR, Bajgai TR, Dhital R, Khanal S, Koirala N, Raghavan V (2020) Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation 6:59. https://doi.org/10.3390/fermentation6020059
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likehood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Thakur JP, Haider R, Singh DK, Balagani SK, Vasudev PG, Luqman S, Kalra A, Saikia D, Negi AS (2015) Bioactive isochromenone isolated from Aspergillus fumigates, endophytic fungus from Bacopa monnieri. Microbiol Res 6:5800
Ververis C, Georghiou K, Danielidis D, Hatzinikolaou DG, Santas P, Santas R, Corleti V (2007) Cellulose, hemicelluloses, lignin and ash content of some organic materials and their suitability for use as paper pulp supplements. Bioresou Technol 98:296–301
Wood TM, Bhat KM (1988) Methods for measuring cellulase activities. Methods Enzymol 160:87–112. https://doi.org/10.1016/0076-6879(88)60109-1
Acknowledgements
The research was funded by AMAAS networking project of Indian Council of Agricultural research, New Delhi, India. The author also wishes to thank Director, ICAR-CISH for providing the necessary facilities during the course of investigation.
Funding
The research was funded by ‘Application of Microorganisms in Agriculture and Allied Sectors’ (AMAAS) networking project of Indian Council of Agricultural research, New Delhi, India.
Author information
Authors and Affiliations
Contributions
NG and PM: conceived the research, designed experiments; BS, SV and SK: performed the experiments; BS, SKS and SK: analyzed the data and MS writing; NG, BS and SKS editing the MS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research content
The research content of manuscript is original and has not been published elsewhere.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, B., Garg, N., Mathur, P. et al. Microbial production of multienzyme preparation from mosambi peel using Trichoderma asperellum. Arch Microbiol 204, 313 (2022). https://doi.org/10.1007/s00203-022-02913-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02913-x