Abstract
A Gram-positive, rod-shaped, spore-forming, thermophilic, and acidophilic bacterium, designated as strain skT53T, was isolated from farm soil in Tokyo, Japan. Under aerobic conditions, the strain grew at 35–55 °C (optimum temperature 44–55 °C) and pH 4.0–6.0 (optimum pH 5.0). Phylogenetic analysis of the 16S rRNA gene sequence showed that the isolate was moderately related to the type strain of Effusibacillus consociatus (94.3% similarity). The G + C content of the genomic DNA was 48.2 mol%, and MK-7 was the predominant respiratory quinone in the strain. The major fatty acids were anteiso-C15:0, iso-C15:0, and iso-C16:0. Based on the phenotypic and chemotaxonomic characteristics, as well as 16S rRNA gene sequence similarity and whole genome analyses, strain skT53T represents a novel species in the genus Effusibacillus, for which the name Effusibacillus dendaii sp. nov. has been proposed. The type strain is skT53T (= NBRC 114101 T = TBRC 11241 T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Alicyclobacillaceae was established by da Costa and Rainey (2009) to accommodate only the genus Alicyclobacillus, which includes spore-forming, acidophilic, and thermophilic bacteria with cellular fatty acids comprising ω-alicyclic fatty acids (Wisotzkey et al. 1992). The genera Kyrpidia (Klenk et al. 2011) and Tumebacillus (Steven et al. 2008) have been added to this family since it was first established. Species belonging to the genus Kyrpidia, which contains two species (Bonjour and Arago 1984; Reiner et al. 2018), are spore-forming, thermophilic, and acidophilic; whereas, species belonging to the genus Tumebacillus are spore-forming and mesophilic. Although typical bacteria belonging to the genus Alicyclobacillus possess ω-alicyclic fatty acids, a few members with no detectable ω-alicyclic fatty acids have also been identified; among them, two species named A. consociatus and A. pohliae, which are phylogenetically distinct from previously known species, have been established (Glaeser et al. 2013; Imperio et al. 2008). Based on 16S rRNA gene sequencing analysis, these two species were subsequently reclassified into the new genus Effusibacillus along with E. lacus in 2014 (Watanabe et al. 2014). In the present study, a novel strain, termed skT53T, isolated from farm soil was taxonomically studied using polyphasic approaches. Herein, we propose a new species of the genus Effusibacillus.
Materials and methods
Sampling and isolation
Strain skT53T was isolated from the soil of a farm in Adachi-ku Tokyo (35 °47 ′01.3 ″ " N, 139 °47 ′58.0 "″ E), where crop failure occurred in 2014.
Briefly, 1 g of soil sample was suspended in 4.5 mL of sterile saline, and the suspension was allowed to stand for 10 min prior to the recovery of the supernatant. The supernatant was serially diluted and plated on SKT medium containing (per liter) 0.8 mg Difco nutrient broth, 6 g gellan gum, and 0.16 g CaCl2. The inoculated plates were incubated at 37 °C for 4 weeks. To select microorganisms that exhibited growth only in oligotrophic media, colonies formed were transferred to both SKT and LB media plates. Microorganisms that showed growth on SKT medium without demonstrating growth on LB medium were selected. Selected microorganisms were suspended in sterile saline and incubated at 80 °C for 30 min to enable the formation of spores, and each suspension was then plated onto SKT medium. After incubation at 37 °C for 4 weeks, growth of nine microorganisms was observed; these were then collected and stored for further analysis. The nine microorganisms were suspended individually in sterile saline, and the cell suspensions were incubated at 80 °C for 30 min to facilitate spore formation. After incubation, sporulation of all nine microorganisms was confirmed using a Wirtz spore staining kit (Muto Pure Chemicals, Japan). The 16S rRNA genes of the nine microorganisms were amplified through PCR with the primers 27f (= 8f) and 1492r (Turner et al. 1999; Loy et al. 2002), and their sequences were analyzed. Among the nine microorganisms, we selected the one with the lowest similarity in 16S rRNA gene sequences compared to those with known sequences and designated it as strain skT53T.
Phenotypic and microscopic analysis and growth conditions
Analysis of cell morphology, assessment using Gram staining, determination of catalase and oxidase activities, and examination of optimal medium conditions were performed by the Identification Service of Techno Suruga Lab Co., Ltd., Japan. Cell morphology was observed using a stereomicroscope (SMZ800N; Nikon, Japan). Gram staining results were observed under a light microscope (BX50F4; Olympus, Japan) using a Gram staining kit (Nissui Pharmaceutical, Japan). Catalase and oxidase activities were determined according to the method described by Barrow and Feltham (1993). To assess anaerobic growth, the growth of the strain skT53 on a yeast extract mineral medium plate (DSM Medium 259, https://www.dsmz.de/collection/catalogue/microorganisms/culture-technology/list-of-media-for-microorganisms) was analyzed using the AnaeroPack System (Mitsubishi Gas Chemical, Japan) (Delaney and Onderdonk 1997).
Each experiment for characterization of strain skT53T was conducted in triplicate at 50 °C (excluding the growth temperature test) using the yeast extract mineral medium with pH adjusted to pH 5.0 (excluding the pH test). To evaluate the optimum growth temperature, cultures were incubated at 11 different temperatures ranging from 30 °C to 80 °C, using liquid media with incubation temperatures ranging from 30 °C to 50 °C as well as solid media with incubation temperatures ranging from 50 °C to 80 °C. To evaluate optimal growth pH, cultures were incubated using solid media with pH adjusted to pH 4.0, 5.0, 6.0, and 7.0, with HCl or NaOH solution according to previously described methods (Sakamoto et al. 2017).
The utilization of organic substrates was assessed using a liquid medium containing (per liter) 1.0 g MgSO4.7H2O, 0.1 g CaCl2.2H2O, 0.1 g NH4Cl, 0.1 g KH2PO4, 0.1 g KCl, 1 mL trace element solution, 1 mL selenite tungstate solution, 1 mL vitamin mixture, 1 mL vitamin B12 solution, and 1 mL thiamine solution (Watanabe et al. 2014), buffered with 20 mM MES-NaOH (pH 5.0) under oxic conditions. All stock solutions were prepared according to the protocol described by Widdel and Bak (1992). Each substrate was added to the defined medium at a final concentration of 10 mM. The substrates tested included carbohydrates (D-glucose, D-galactose, D-arabinose, sucrose, cellobiose, D-fructose, maltose, mannose, melibiose, D-sorbitol, trehalose, D-xylose, and N-acetylglucosamine) and organic acids (acetate, fumarate, D-lactate, L-lactate, and succinate). Additionally, yeast extract was tested at a final concentration of 2 g/L. The sugar oxidation test was performed using an API50CH (BioMérieux, France). The enzyme activity test was performed using APIZYM (BioMérieux, France).
Chemotaxonomic analysis
Menaquinone extraction was performed according to the methods described by Collins et al. (1977), and analysis was performed using HPLC (Kroppenstedt 1982). Polar lipids were extracted from 100 mg of freeze-dried cells, purified using the methods described by Minnikin et al. (1979), and analyzed via thin-layer chromatography using chloroform/methanol/water (65: 25: 4, by volume) in the first direction and chloroform/acetic acid/methanol/water (80: 18: 12: 5, by volume) in the second. Cellular fatty acid methyl esters were identified and quantified by gas chromatography (6890 N; Agilent Technologies, USA) according to the standard protocol of the Sherlock Microbial Identification System (Sasser 1990) with the Sherlock Midi software (version 6.2) and the TSBA6 database. Amino acids of peptidoglycans were analyzed as described previously (Hamada et al. 2012). The isomer of diaminopimelic acid (DAP) in the cell wall peptidoglycan was determined as described by Hasegawa (1983).
The 16S rRNA gene sequencing and phylogenetic analysis
The 16S rRNA gene fragment was amplified using the universal primers 9F and 1510R. The nucleotide sequence of the amplified fragment was determined by Fasmac Co., Ltd. (Japan), using the primers 9F, 515F, 1099F, 536R, 926R, and 1510R (Lane et al. 1985; Turner et al. 1999; Lane 1991; Nakagawa et al. 2001). The almost-complete 16S rRNA gene sequence (1471 nt) of strain skT53T was compared with that of the type strains of species with valid published names using EzBioCloud (Yoon et al. 2017a). The CLUSTAL X program (Thompson et al. 1997) was used to align the 16S rRNA gene sequence of strain skT53T with the corresponding sequences of the family Alicyclobacillaceae. Phylogenetic trees were reconstructed using the neighbor-joining (NJ) (Saitou et al. 1987), maximum-likelihood (ML) (Felsenstein 1981), and maximum-parsimony (MP) (Fitch 1971) algorithms using the MEGA X program (Kumar et al. 2018). The resultant tree topologies were evaluated by performing bootstrap analysis (Felsenstein 1985) based on 1000 replicates.
Genome sequencing and analysis
Genomic DNA extraction of the strain skT53T was conducted using cultured cells and the Wizard® Genomic DNA Purification Kit (Promega, USA). Genome sequencing was performed by Macrogen Japan Co., Ltd. (Japan) using the PacBio RSII. The reads of each strain were assembled using the FALCON-integrated version 2.14. The DNA G + C content of strain skT53T was 48.2 mol%. The consensus phylogenetic tree was constructed based on the data of a multi-locus alignment of core genes in the strain skT53T with related species in the NCBI Assembly database using the automated multi-locus species tree (autoMLST) (https://automlst.ziemertlab.com) (Alanjary et al. 2019).
The average nucleotide identity (ANI) and the digital DNA–DNA hybridization (dDDH) values were used to calculate genomic similarities between strain skT53T and the type strains of other Effusibacillus species. The ANI based on BLAST was determined using the ANI Calculator (https://www.ezbiocloud.net/tools/ani) (Yoon et al. 2017b; Goris et al. 2007), and the dDDH values were calculated using the Genome-to-Genome Distance Calculator 2.1 (GGDC; http://ggdc.dsmz.de/distcalc2.php) (Auch et al. 2010; Meier-Kolthoff et al. 2013a, b). Formula 2 was applied to dDDH analysis.
Results and discussion
The cells of strain skT53T were rod shaped, and Gram staining results indicated that the cells were Gram positive. The diameter of the cells grown on yeast extract mineral medium ranged from 0.6 to 0.8 µm, and their length ranged from 2 to 10 µm. Spores were observed at high temperatures. The phenotypic characteristics of the strain skT53T are shown in Table 1.
The skT53T strain did not grow with the sugars tested but grew aerobically on yeast extract, acetate, fumarate, D-lactate and succinate. No sugar was oxidized in the sugar oxidation test using API50CH. The strain showed weak anaerobic growth under the conditions described in the Materials and methods. Evaluation of enzyme activity using APIZYM revealed that the strain skT53T exhibits alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine allyl amidase, acid phosphatase, and naphthol-AS-BI-phosphohydrase activities.
The major cellular fatty acids (> 10% of the total) of strain skT53T were anteiso-C15:0 (18.26%), iso-C15:0 (17.40%), and iso-C16:0 (15.36%); the total cell fatty acid profile of the strain is shown in Table 2. The respiratory quinones in the strain were MK-7 (97.5%) and MK-8 (2.4%). Polar lipids included diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylmethylethanolamine, three unidentified phospholipids, and two unidentified polar lipids. The peptidoglycan sample contained alanine, glutamic acid, and meso-diaminopimelic acid. All chemotaxonomic data are consistent with the description of the genus Effusibacillus.
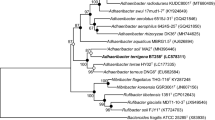
The 16S rRNA gene sequence indicated that the species most closely related to strain skT53T were E. consociatus CCUG53762T strain (94.3%; Glaeser et al. 2013), E. lacus skLN1T (93.4%; Watanabe et al. 2014) and E. pohliae MP4 (93.5%; Imperio et al. 2008). Strain skT53T showed less than 91.3% 16S rRNA gene sequence similarity with other members of the family Alicyclobacillaceae. These results are also in line with the classification of strain skT53T in the genus Effusibacillus. Phylogenetic analysis based on the 16S rRNA gene sequences revealed that strain skT53T forms a clade with members of the genus Effusibacillus supported by a high bootstrap value (Fig. 1) and ML and MP algorithms (Supplemental Fig. S1 and S2). As shown in Fig. 1 (NJ), strain skT53T formed a clade with members of the genus Effusibacillus, and the genera Alicyclobacillus, Tumebacillus, and Effusibacillus formed independent clades. In Supplemental Fig. S1 (ML) and S2 (MP), strain skT53T formed a clade with members of the genera Effusibacillus and Tumebacillus. Members of the genus Tumebacillus formed a daughter clade against the genus Effusibacillus. These results show that strain skT53T phylogenetically belongs to the genus Effusibacillus. In addition, these results based on the 16S rRNA gene sequences were consistent with the phylogenetic analysis based on the multi-locus alignment of core genes (Fig. 2).
Phylogenetic tree based on 16S rRNA gene sequences created using the neighbor-joining method in MEGA X (Kumar et al. 2018), showing the position of strain skT53T and type strains within the family Alicyclobacillaceae. Numbers at branching points refer to percentages of bootstrap values over 50% derived from 1000 replications. Bar, 0.01 substitutions per nucleotide position
The consensus phylogenetic tree was built from a multi-locus alignment of core genes in strain skT53T with Alicyclobacillaceae species in the NCBI Assembly database by using the automated multi-locus species tree (autoMLST) (https://automlst.ziemertlab.com)
Strain skT53T exhibited dDDH values of 22.5% for E. lacus DSM 27172 T and 18.8% for E. pohliae DSM 22757 T; both values are clearly below the 70% threshold for the definition of bacterial species (Wayne et al. 1987). Strain skT53T exhibited an ANI of 72.0% for E. pohliae DSM 22757 T and 71.8% for E. lacus DSM 27172 T. These results are significantly below 95–96% (Goris et al. 2007) recommended cut-off points for ANI values.
Based on the data presented, we conclude that strain skT53T represents a novel species of the genus Effusibacillus, for which the name E. dendaii sp. nov. is proposed.
Description of Effusibacillus dendaii sp. nov.
Effusibacillus dendaii (den.dai’i. N.L. gen. masc. n., Dendai’s dendaii (the Japanese abbreviation name of Tokyo Denki University, where this isolate was initially characterized).
The isolate is a facultatively anaerobic, gram-positive, spore-forming, and rod-shaped bacterium with a cell size of 0.6–0.8 × 2–10 µm. The temperature range for growth is 35–55 °C, and the optimum growth temperature is 44–55 °C. The pH range for growth is 4.0–6.0, and the optimum pH was 5.0. It is oxidase positive but catalase negative. It utilizes yeast extract, acetate, fumarate, D-lactate, and succinate as carbon sources. The major fatty acids are anteiso-C15:0, iso-C15:0, and iso-C16:0. The major respiratory quinone is MK-7. The polar lipids comprise diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylmethylethanolamine, three unidentified phospholipids, and two unidentified polar lipids. The diagnostic diamino acid in the cell wall peptidoglycan is meso-DAP. The in silico genomic DNA G + C content of the type strain is 48.2 mol%.
The type strain, skT53T (NBRC 114101 T = TBRC 11241 T), was isolated from farm soil in Adachi-ku, Tokyo, Japan.
References
Alanjary M, Steinke K, Ziemert N (2019) AutoMLST: an automated web server for generating multi-locus species trees highlighting natural product potential. Nuc Acids Res 47:W276–W282. https://doi.org/10.1093/nar/gkz282
Auch AF, von Jan M, Klenk HP, Göker M (2010) Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2:117–134. https://doi.org/10.4056/sigs.531120
Barrow GI, Feltham RKA (1993) Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd edn. University Press, Cambridge
Bonjour F, Aragno M (1984) Bacillus tusciae, a new species of thermoacidophilic, facultatively chemolithoautotrophic, hydrogen oxidizing sporeformer from a geothermal area. Arch Microbiol 139:397–401. https://doi.org/10.1007/BF00408386
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
da Costa MS, Rainey FA (2009) Family II Alicyclobacillaceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol 3, 2nd edn. Springer, New York
Delaney ML, Onderdonk AB (1997) Evaluation of the AnaeroPack system for growth of clinically significant anaerobes. J Clin Microbiold 35:558–562. https://doi.org/10.1128/jcm.35.3.558-562.1997
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Glaeser SP, Falsen E, Martin K, Kämpfer P (2013) Alicyclobacillus consociatus sp. nov., isolated from a human clinical specimen. Int J Syst Evol Microbiol 63:3623–3627. https://doi.org/10.1099/ijs.0.048173-0
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P et al (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Hamada M, Yamamura H, Komukai C, Tamura T, Suzuki K et al (2012) Luteimicrobium album sp. nov., a novel actinobacterium isolated from a lichen collected in Japan, and emended description of the genus Luteimicrobium. J Antibiot 65:427–431. https://doi.org/10.1038/ja.2012.45
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322. https://doi.org/10.2323/jgam.29.319
Imperio T, Viti C, Marri L (2008) Alicyclobacillus pohliae sp. nov., a thermophilic, endospore-forming bacterium isolated from geothermal soil of the north-west slope of Mount Melbourne (Antarctica). Int J Syst Evol Microbiol 58:221–225. https://doi.org/10.1099/ijs.0.65092-0
Klenk HP, Lapidus A, Chertkov O, Copeland A, Del Rio TG et al (2011) Complete genome sequence of the thermophilic, hydrogen-oxidizing Bacillus tusciae type strain (T2) and reclassification in the new genus, Kyrpidia gen. nov. as Kyrpidia tusciae comb. nov. and emendation of the family Alicyclobacillaceae da Costa and Rainey, 2010. Stand Genomic Sci 5:121–134. https://doi.org/10.4056/sigs.2144922
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP 18) and a silver loaded ion exchanger as stationary phases. J Liquid Chromatogr 5:2359–2387. https://doi.org/10.1080/01483918208067640
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lane DJ (1991) 16S/23S rDNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics, J. Wiley & Sons, Chicester, England, pp 115–175
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML et al (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82:6955–6959. https://doi.org/10.1073/pnas.82.20.6955
Loy A, Lehner A, Lee N, Adamczyk J, Meier H et al (2002) Oligonucleotide microarray for 16s rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081. https://doi.org/10.1128/AEM.68.10.5064-5081.2002
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013a) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Meier-Kolthoff JP, Göker M, Spröer C, Klenk HP (2013b) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418. https://doi.org/10.1007/s00203-013-0888-4
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95. https://doi.org/10.1111/j.1365-2672.1979.tb01172.x
Nakagawa Y, Kawasaki H (2001) Methods in gene analysis. In: Miyadoh S, Hamada M, Hotta K, Kudo T, Seino A, Suzuki K, Yokota A (eds) Identification manual of actinomycetes. Business center for academic societies Japan, Tokyo, pp 88–117
Reiner JE, Jung T, Lapp CJ, Siedler M, Bunk B et al (2018) Kyrpidia spormannii sp. nov., a thermophilic, hydrogen-oxidizing, facultative autotroph, isolated from hydrothermal systems at Sao Miguel Island, and emended description of the genus Kyrpidia. Int J Syst Evol Microbiol 68:3735–3740. https://doi.org/10.1099/ijsem.0.003037
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sakamoto M, Iino T, Ohkuma M (2017) Faecalimonas umbilicata gen. nov., sp. nov., isolated from human faeces, and reclassification of Eubacterium contortum, Eubacterium fissicatena and Clostridium oroticum as Faecalicatena contorta gen. nov., comb. nov., Faecalicatena fissicatena comb. nov. and Faecalicatena orotica comb. nov. Int J Syst Evol Microbiol 67:1219–1227. https://doi.org/10.1099/ijsem.0.001790
Sasser M (1990) Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. MIDI Inc, Newark, DE
Steven B, Chen MQ, Greer CW, Whyte LG, Niederberger TD (2008) Tumebacillus permanentifrigoris gen. nov., sp. nov., an aerobic, spore-forming bacterium isolated from Canadian high Arctic permafrost. Int J Syst Evol Microbiol 58:1497–1501. https://doi.org/10.1099/ijs.0.65101-0
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Turner S, Pryer KM, Miao VPW, Palmer JD (1999) Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46:327–338. https://doi.org/10.1111/j.1550-7408.1999.tb04612.x
Watanabe M, Kojima H, Fukui M (2014) Proposal of Effusibacillus lacus gen. nov., sp. nov., and reclassification of Alicyclobacillus pohliae as Effusibacillus pohliae comb. nov. and Alicyclobacillus consociatus as Effusibacillus consociatus comb. nov. Int J Syst Evol Microbiol 64:2770–2774. https://doi.org/10.1099/ijs.0.055814-0
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O et al (1987) Report of the AD hoc Committee on reconciliation of approaches to bacterial Systematics. Int J Syst Evol Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate reducing bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The Prokaryotes, vol 4, 2nd edn. Springer, New York, pp 3352–3378
Wisotzkey JD, Jurtshuk P Jr, Fox GE, Deinhard G, Poralla K (1992) Comparative sequence analyses on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. nov. Int J Syst Bacteriol 42:263–269. https://doi.org/10.1099/00207713-42-2-263
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y et al (2017a) Introducing EzBioCloud: a taxonomically United database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Acknowledgements
We would like to thank Mr. Kiyohide Ushigome, a farmer in Adachi-ku, for providing us with farm soil. We would like to thank Editage (www.editage.com) for English language editing.
Funding
This research was supported in part by a grant-in-aid from the Institute for Fermentation, Osaka, to H.K. and the environment fund from Adachi-ku (No. 27–1363) to H.K.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the genome sequence of strain skT53 is AP023366. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain skT53 is LC586281.
Supplementary Information
Below is the link to the electronic supplementary material.
203_2021_2470_MOESM1_ESM.pptx
Supplementary file1 (PPTX 69 KB) Phylogenetic tree based on 16S rRNA gene sequences created using the maximum-likelihood method in MEGA X (Kumar et al., 2018), showing the phylogenetic positions of strain skT53T and type strains within the family Alicyclobacillaceae. Numbers at branching points refer to percentages of bootstrap values over 50% derived from 1000 replications. Bar, 0.02 substitutions per nucleotide position.
203_2021_2470_MOESM2_ESM.pptx
Supplementary file2 (PPTX 69 KB) Phylogenetic tree based on 16S rRNA gene sequences created using the maximum-parsimony method in MEGA X (Kumar et al., 2018), showing the phylogenetic positions of strain skT53T and type strains within the family Alicyclobacillaceae. Numbers at branching points refer to percentages of bootstrap values over 50% derived from 1000 replications.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Konishi, T., Tamura, T., Tobita, T. et al. Effusibacillus dendaii sp. nov. isolated from farm soil. Arch Microbiol 203, 4859–4865 (2021). https://doi.org/10.1007/s00203-021-02470-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02470-9