Abstract
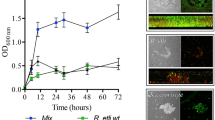
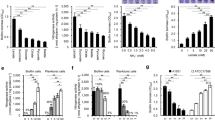
Organisms belonging to the genus Rhizobium colonize leguminous plant roots and establish a mutually beneficial symbiosis. Biofilms are structured ecosystems in which microbes are embedded in a matrix of extracellular polymeric substances, and their development is a multistep process. The biofilm formation processes of R. etli CFN42 were analyzed at an early (24-h incubation) and mature stage (72 h), comparing cells in the biofilm with cells remaining in the planktonic stage. A genome-wide microarray analysis identified 498 differentially regulated genes, implying that expression of ~8.3 % of the total R. etli gene content was altered during biofilm formation. In biofilms-attached cells, genes encoding proteins with diverse functions were overexpressed including genes involved in membrane synthesis, transport and chemotaxis, repression of flagellin synthesis, as well as surface components (particularly exopolysaccharides and lipopolysaccharides), in combination with the presence of activators or stimulators of N-acyl-homoserine lactone synthesis This suggests that R. etli is able to sense surrounding environmental conditions and accordingly regulate the transition from planktonic and biofilm growth. In contrast, planktonic cells differentially expressed genes associated with transport, motility (flagellar and twitching) and inhibition of exopolysaccharide synthesis. To our knowledge, this is the first report of nodulation and nitrogen assimilation-related genes being involved in biofilm formation in R. etli. These results contribute to the understanding of the physiological changes involved in biofilm formation by bacteria.





Similar content being viewed by others
References
Anderson PE, Gober JW (2000) FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol Microbiol 38:41–52
Andrade-Dominguez A, Salazar E, Vargas-Lagunas Mdel C et al (2014) Eco-evolutionary feedbacks drive species interactions. ISME J 8:1041–1054. doi:10.1038/ismej.2013.208
Ausmees N, Jacobsson K, Lindberg M (2001) A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology 147:549–559
Bayles KW (2007) The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5:721–726. doi:10.1038/nrmicro1743
Belas R, Horikawa E, Aizawa SI, Suvanasuthi R (2009) Genetic determinants of Silicibacter sp. TM1040 motility. J Bacteriol 191:4502–4512. doi:10.1128/JB.00429-09
Belotserkovsky JM, Dabbs ER, Isaksson LA (2011) Mutations in 16S rRNA that suppress cold-sensitive initiation factor 1 affect ribosomal subunit association. FEBS J 278:3508–3517. doi:10.1111/j.1742-4658.2011.08272.x
Celano B, Pawlik RT, Gualerzi CO (1988) Interaction of Escherichia coli translation-initiation factor IF-1 with ribosomes. Eur J Biochem 178:351–355
Christensen LD, Moser C, Jensen PO et al (2007) Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153:2312–2320. doi:10.1099/mic.0.2007/006122-0
Chua SL, Hultqvist LD, Yuan M et al (2015) In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat Protoc 10:1165–1180. doi:10.1038/nprot.2015.067
Danhorn T, Fuqua C (2007) Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi:10.1146/annurev.micro.61.080706.093316
Daniels R, Reynaert S, Hoekstra H et al (2006) Quorum signal molecules as biosurfactants affecting swarming in Rhizobium etli. Proc Natl Acad Sci USA 103:14965–14970. doi:10.1073/pnas.0511037103
Dardanelli M, Angelini J, Fabra A (2003) A calcium-dependent bacterial surface protein is involved in the attachment of rhizobia to peanut roots. Can J Microbiol 49:399–405. doi:10.1139/w03-054
Davey ME, O’Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867
de Lucena DK, Puhler A, Weidner S (2010) The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol 10:265. doi:10.1186/1471-2180-10-265
de Vries S, Hoge H, Bisseling T (1989) Isolation of total and polysomal RNA from plant tissues. Springer, Netherlands
Donlan RM (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890
Dressaire C, Moreira RN, Barahona S et al (2015) BolA is a transcriptional switch that turns off motility and turns on biofilm development. MBio 6:e02352-14. doi:10.1128/mBio.02352-14
Flechard M, Fontenelle C, Blanco C et al (2010) RpoE2 of Sinorhizobium meliloti is necessary for trehalose synthesis and growth in hyperosmotic media. Microbiology 156:1708–1718. doi:10.1099/mic.0.034850-0
Fraysse N, Couderc F, Poinsot V (2003) Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270:1365–1380
Frederix M, Edwards A, Swiderska A et al (2014) Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol Microbiol 93:464–478. doi:10.1111/mmi.12670
Fujishige NA, Rinaudi L, Giordano W, Hirsch A (2005) Superficial liaisons: colonization of roots and abiotic surfaces by rhizobia. In: Sánchez F, Quinto C, López-Lara IM, Geiger O (eds) Biology of Plant-Microbe Interactions, Proceedings of the 12th International Congress on Molecular Plant–Microbe Interactions, St. Paul, MN, pp 292–299
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56:195–206. doi:10.1111/j.1574-6941.2005.00044.x
Fujishige NA, Lum MR, De Hoff PL et al (2008) Rhizobium common nod genes are required for biofilm formation. Mol Microbiol 67:504–515. doi:10.1111/j.1365-2958.2007.06064.x
Fuqua WC, Winans SC (1994) A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806
Gardner SG, Johns KD, Tanner R, McCleary WR (2014) The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol 196:1741–1752. doi:10.1128/JB.00029-14
Hegde P, Qi R, Abernathy K et al (2000) A concise guide to cDNA microarray analysis. Biotechniques 29:548–50, 552–4, 556 passim
Israel DW (1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84:835–840
Jafri S, Urbanowski ML, Stauffer GV (1995) A mutation in the rpoA gene encoding the alpha subunit of RNA polymerase that affects metE-metR transcription in Escherichia coli. J Bacteriol 177:524–529
Javaherdashti R, Nwaoha C, Tan H (2013) Corrosion and materials in the oil and gas industries. CRC Press, Boca Raton
Jayasinghearachchi HS, Seneviratne G (2006) Fungal solubilization of rock phosphate is enhanced by forming fungal–rhizobial biofilms. Soil Biol Biochem 38:405–408. doi:10.1016/j.soilbio.2005.06.004
Jensen JB, Peters NK, Bhuvaneswari TV (2002) Redundancy in periplasmic binding protein-dependent transport systems for trehalose, sucrose, and maltose in Sinorhizobium meliloti. J Bacteriol 184:2978–2986
Jitacksorn S, Sadowsky MJ (2008) Nodulation gene regulation and quorum sensing control density-dependent suppression and restriction of nodulation in the Bradyrhizobium japonicum-soybean symbiosis. Appl Environ Microbiol 74:3749–3756. doi:10.1128/AEM.02939-07
Kamath S, Kapatral V, Chakrabarty AM (1998) Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol 30:933–941
Karatan E, Watnick P (2009) Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi:10.1128/MMBR.00041-08
Kim HS, Park SJ, Lee KH (2009) Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol 74:436–453. doi:10.1111/j.1365-2958.2009.06875.x
Ko JH, Lee SJ, Cho B, Lee Y (2006) Differential promoter usage of infA in response to cold shock in Escherichia coli. FEBS Lett 580:539–544. doi:10.1016/j.febslet.2005.12.066
Kusada H, Hanada S, Kamagata Y, Kimura N (2014) The effects of N-acylhomoserine lactones, beta-lactam antibiotics and adenosine on biofilm formation in the multi-beta-lactam antibiotic-resistant bacterium Acidovorax sp. strain MR-S7. J Biosci Bioeng 118:14–19. doi:10.1016/j.jbiosc.2013.12.012
Laus MC, Logman TJ, Van Brussel AA et al (2004) Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 186:6617–6625. doi:10.1128/JB.186.19.6617-6625.2004
Leigh JA, Dodsworth JA (2007) Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol 61:349–377. doi:10.1146/annurev.micro.61.080706.093409
Lerouge P, Roche P, Faucher C et al (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344:781–784. doi:10.1038/344781a0
Locke AE, Kahali B, Berndt SI et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206. doi:10.1038/nature14177
Loh J, Garcia M, Stacey G (1997) NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol 179:3013–3020
Martinez-Salazar JM, Salazar E, Encarnacion S et al (2009a) Role of the extracytoplasmic function sigma factor RpoE4 in oxidative and osmotic stress responses in Rhizobium etli. J Bacteriol 191:4122–4132. doi:10.1128/JB.01626-08
Martinez-Salazar JM, Sandoval-Calderon M, Guo X et al (2009b) The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155:386–397. doi:10.1099/mic.0.021428-0
Metselaar KI, Saa Ibusquiza P, Ortiz Camargo AR et al (2015) Performance of stress resistant variants of Listeria monocytogenes in mixed species biofilms with Lactobacillus plantarum. Int J Food Microbiol. doi:10.1016/j.ijfoodmicro.2015.04.021
Modarresi F, Azizi O, Shakibaie MR et al (2015) Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 6:152–161. doi:10.1080/21505594.2014.1003001
Mongiardini EJ, Ausmees N, Pérez-Giménez J et al (2008) The rhizobial adhesion protein RapA1 is involved in adsorption of rhizobia to plant roots but not in nodulation. FEMS Microbiol Ecol 65:279–288. doi:10.1111/j.1574-6941.2008.00467.x
Numata T, Ikeuchi Y, Fukai S et al (2006) Crystallization and preliminary X-ray analysis of the tRNA thiolation enzyme MnmA from Escherichia coli complexed with tRNA(Glu). Acta Crystallogr Crystallogr F Struct Biol Cryst Commun 62:368–371. doi:10.1107/S174430910600738X
O’Toole GA, Kolter R (1998a) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
O’Toole GA, Kolter R (1998b) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461
Oh MH, Choi CH (2015) Role of LuxIR Homologue AnoIR in Acinetobacter nosocomialis and the Effect of Virstatin on the Expression of anoR Gene. J Microbiol Biotechnol. doi:10.4014/jmb.1504.04069
Ottemann KM, Miller JF (1997) Roles for motility in bacterial-host interactions. Mol Microbiol 24:1109–1117
Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293
Qin Y, Luo ZQ, Farrand SK (2004) Domains formed within the N-terminal region of the quorum-sensing activator TraR are required for transcriptional activation and direct interaction with RpoA from agrobacterium. J Biol Chem 279:40844–40851. doi:10.1074/jbc.M405299200M405299200
Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32(Suppl):496–501. doi:10.1038/ng1032ng1032
Ramos JL, Martinez-Bueno M, Molina-Henares AJ et al (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi:10.1128/MMBR.69.2.326-356.2005
Rasamiravaka T, Labtani Q, Duez P, El Jaziri M (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015:759348. doi:10.1155/2015/759348
Ribeiro RA, Barcellos FG, Thompson FL, Hungria M (2009) Multilocus sequence analysis of Brazilian Rhizobium microsymbionts of common bean (Phaseolus vulgaris L.) reveals unexpected taxonomic diversity. Res Microbiol 160:297–306. doi:10.1016/j.resmic.2009.03.009
Rinaudi LV, Giordano W (2010) An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett 304:1–11. doi:10.1111/j.1574-6968.2009.01840.x
Robleto EA, Kmiecik K, Oplinger ES et al (1998) Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl Environ Microbiol 64:2630–2633
Russo DM, Williams A, Edwards A et al (2006) Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J Bacteriol 188:4474–4486. doi:10.1128/JB.00246-06
Sadovskaya I, Vinogradov E, Li J et al (2010) High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1- > 3)-glucans, which bind aminoglycosides. Glycobiology 20:895–904. doi:10.1093/glycob/cwq047
Salazar E, Diaz-Mejia JJ, Moreno-Hagelsieb G et al (2010) Characterization of the NifA-RpoN regulon in Rhizobium etli in free life and in symbiosis with Phaseolus vulgaris. Appl Environ Microbiol 76:4510–4520. doi:10.1128/AEM.02007-09
Schmidt J, Musken M, Becker T et al (2011) The Pseudomonas aeruginosa chemotaxis methyltransferase CheR1 impacts on bacterial surface sampling. PLoS One 6:e18184. doi:10.1371/journal.pone.0018184
Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32:33–57. doi:10.1146/annurev.genet.32.1.33
Smith DR, Chapman MR (2010) Economical evolution: microbes reduce the synthetic cost of extracellular proteins. MBio. doi:10.1128/mBio.00131-10
Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100:9440–9445. doi:10.1073/pnas.15305091001530509100
Tatusov RL, Fedorova ND, Jackson JD et al (2003) The COG database: an updated version includes eukaryotes. BMC Bioinform 4:41. doi:10.1186/1471-2105-4-411471-2105-4-41
Vorachit M, Lam K, Jayanetra P, Costerton JW (1995) Electron microscopy study of the mode of growth of Pseudomonas pseudomallei in vitro and in vivo. J Trop Med Hyg 98:379–391
Wang P, Zhong Z, Zhou J et al (2008) Exopolysaccharide biosynthesis is important for Mesorhizobium tianshanense: plant host interaction. Arch Microbiol 189:525–530. doi:10.1007/s00203-007-0345-3
Workman C, Jensen LJ, Jarmer H et al (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 3. doi:10.1186/gb-2002-3-9-research0048.1
Yeom S, Yeom J, Park W (2010) NtrC-sensed nitrogen availability is important for oxidative stress defense in Pseudomonas putida KT2440. J Microbiol 48:153–159. doi:10.1007/s12275-010-0075-0
Young JPW, Crossman LC, Johnston AWB et al (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34. doi:10.1186/gb-2006-7-4-r34
Zarkani AA, Stein E, Rohrich CR et al (2013) Homoserine lactones influence the reaction of plants to rhizobia. Int J Mol Sci 14:17122–17146. doi:10.3390/ijms140817122
Zhang Y, Pohlmann EL, Roberts GP (2005) GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J Bacteriol 187:1254–1265. doi:10.1128/JB.187.4.1254-1265.2005
Zheng H, Zhong Z, Lai X et al (2006) A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188:1943–1949. doi:10.1128/JB.188.5.1943-1949.2006
Zheng H, Mao Y, Zhu Q et al (2015) The quorum sensing regulator CinR hierarchically regulates two other quorum sensing pathways in ligand-dependent and-independent fashions in Rhizobium etli. J Bacteriol 197:1573–1581. doi:10.1128/JB.00003-15
Acknowledgments
Part of this work was supported by CONACyT Grant 220790 and DGAPA-PAPIIT Grant IN213216. We are grateful to the Posgrado en Ciencias Biológicas de la Facultad de Ciencias, Universidad Nacional Autónoma de México, México; ARP was a recipient of a PhD Studentship from CONACyT. We thanks to Mario Ramírez Yáñez and Victor Bustos Zagal for technical assistance, to Rubén Paul Gaytán Colín and Eugenio López Bustos from Unidad de Síntesis, Instituto de Biotecnología-UNAM, and to Andrés Saraleguí Amaro and Xochitl Alvarado Affantranger from Unidad Laboratorio Nacional de Microscopía Avanzada-UNAM. Thanks also to Michael Dunn for comments on the manuscript and Enrique Reynaud and Veronica Narvaez for technical assistance in qPCR experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reyes-Pérez, A., Vargas, M., Hernández, M. et al. Transcriptomic analysis of the process of biofilm formation in Rhizobium etli CFN42. Arch Microbiol 198, 847–860 (2016). https://doi.org/10.1007/s00203-016-1241-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1241-5