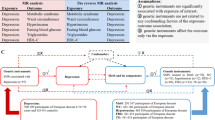
Abstract
Aims/hypothesis
Observational studies have shown a bidirectional association between major depressive disorder (MDD) and cardiometabolic diseases. We conducted a two-sample bidirectional Mendelian randomisation (MR) study to assess the causal associations of MDD with type 2 diabetes, coronary artery disease (CAD) and heart failure and vice versa.
Methods
We extracted summary-level data for MDD, type 2 diabetes, CAD and heart failure from corresponding published large genome-wide association studies of individuals mainly of European-descent. In total, 96 SNPs for MDD, 202 SNPs for type 2 diabetes, 44 SNPs for CAD and 12 SNPs for heart failure were proposed as instrumental variables at the genome-wide significance level (p < 5 × 10−8). The random-effects inverse-variance weighted method was used for the main analyses.
Results
Genetic liability to MDD was significantly associated with type 2 diabetes and CAD at the Bonferroni-corrected significance level. The ORs of type 2 diabetes and CAD were respectively 1.26 (95% CI 1.10, 1.43; p = 6 × 10−4) and 1.16 (95% CI 1.05, 1.29; p = 0.0047) per one-unit increase in loge odds of MDD. There was a suggestive association between MDD and heart failure (OR 1.11 [95% CI 1.01, 1.21]; p = 0.033). We found limited evidence supporting causal effects of cardiometabolic diseases on MDD risk in the reverse MR analyses.
Conclusions/interpretation
The present study strengthened the evidence that MDD is a potential risk factor for type 2 diabetes and CAD. Whether MDD is causally related to heart failure needs further study.
Data availability
All data included in this study were uploaded as supplements and are also publicly available through published GWASs and open GWAS datasets (UK Biobank, 23andMe and Psychiatric Genomics: https://datashare.is.ed.ac.uk/handle/10283/3203; DIAGRAM: http://diagram-consortium.org/downloads.html; CARDIoGRAMplusCD4: www.cardiogramplusc4d.org/; HERMES: http://www.kp4cd.org/datasets/mi).

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.

Introduction
Major depressive disorder (MDD) and type 2 diabetes are both important public health issues globally. It is estimated that around 322 million people had MDD and more than 425 million adults were living with diabetes worldwide in 2015 [1, 2]. Epidemiological data showed that the prevalence of MDD among individuals with type 2 diabetes was twice that in those without diabetes [3], which indicated a potential link between these two prevailing diseases.
Observational studies have reported that MDD is associated with an increased risk of type 2 diabetes through several biological alterations and unhealthy behaviours [4, 5]. It has further been suggested that type 2 diabetes may play a role in the development of MDD, possibly by causing disability and comorbidity [6]. However, whether the mutual association between MDD and type 2 diabetes is causal remains unclear due to potential residual confounding and reverse causation bias in observational studies [7].
Mendelian randomisation (MR) is a method for assessing causal inference of an exposure on an outcome by using genetic variants as instrumental variables for the exposure [8]. This technique diminishes residual confounding because genetic variants are randomly assorted at conception, thereby having no connection to self-selected lifestyle factors, behaviours and environmental factors. In addition, it overcomes reverse causality because genetic variants are fixed regardless of the development or progression of the disease (except for certain cancers).
MDD has been proposed as a risk factor also for other cardiometabolic diseases, such as coronary artery disease (CAD) and heart failure, in observational studies [9,10,11,12]. Therefore, we conducted a bidirectional two-sample MR study to explore the causal associations of MDD with major cardiometabolic diseases as well as the causal role of cardiometabolic diseases for MDD.
Methods
Study design overview
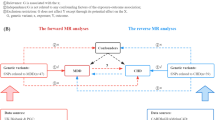
A brief description of the bidirectional MR design is displayed in Fig. 1. There are three key assumptions for MR: (1) genetic variants associate with the exposure of interest; (2) genetic variants are not associated with any confounders of the exposure–outcome association; and (3) genetic variants exert effects on the outcome only via the exposure [13]. We used summary-level data from meta-analyses of genome-wide association studies (GWASs) of MDD, type 2 diabetes, CAD and heart failure. Details of the data sources utilised in the present study are summarised in electronic supplementary material (ESM) Table 1. Studies included in the original GWASs had been approved by a relevant institutional review board and the present MR study has been approved by the Swedish Ethical Review Authority.
Assumptions and study design of the present bidirectional MR study of the associations of MDD with type 2 diabetes, CAD and heart failure. Grey lines show the relationship across instrumental variables, exposure, and outcomes in the MR study examining the effects of MDD on cardiometabolic diseases, and red lines show these relationships in the reverse MR study. Solid lines represent relationship that were observed, whereas dashed lines represent associations that would violate the MR assumptions (i.e. relationships that are not allowed/did not exist in the present MR study). Blue ticks indicate that genetic variants are associated with the exposure/outcome; red crosses indicate that genetic variants are not associated with any confounders of the exposure–outcome association; purple crosses indicate that genetic variants exert effects on the outcome only via the exposure. HF, heart failure; T2DM, type 2 diabetes
Data sources and SNP selection for MDD
We used the hitherto largest published GWAS for MDD that was conducted in a population of European-descent [14], which included 807,553 individuals (246,363 with MDD and 561,190 without MDD) in the discovery stage and 1,306,354 individuals (414,055 with MDD and 892,299 without MDD) in the replication stage [14]. In total, 102 independent variants were identified, of which 96 reached genome-wide significance (p < 5 × 10−8) and were selected as instrumental variables for MDD. Summary-level data from the UK Biobank and Psychiatric Genomics Consortium, including 500,199 individuals (170,756 with MDD and 329,443 without MDD), were publicly available and used in the reverse-direction MR analysis.
Data sources and SNP selection for cardiometabolic diseases
Summary-level data for type 2 diabetes were available from a meta-analysis of 32 GWASs, including 898,130 individuals of European-descent (74,124 with type 2 diabetes and 824,006 without type 2 diabetes) [15]. We selected the 202 SNPs associated with type 2 diabetes at the genome-wide significant level (p < 5 × 10−8) in the model without adjustment for BMI out of 403 distinct association signals. All 403 SNPs, and SNPs associated with type 2 diabetes at p < 5 × 10−8 in the BMI-adjusted model, were used in sensitivity analyses.
Summary-level data for CAD were obtained from an 1000 genomes-based GWAS meta-analysis of 48 studies involving 60,801 individuals with CAD and 123,504 without CAD (77% of participants were of European ancestry) [16]. Fifty-eight distinct loci associated with CAD were discovered in this GWAS, of which 44 SNPs reached genome-wide significance and were used as instrumental variables for CAD in the reverse-direction MR analysis.
Summary-level data for heart failure were extracted from a GWAS of 47,309 individuals with heart failure and 930,014 individuals without heart failure from the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) Consortium [17]. Twelve SNPs were reported to be associated with heart failure at the genome-wide significance level. We included all 12 SNPs in the reverse-direction MR analysis.
Details of the SNPs related to MDD and their associations with cardiometabolic diseases are shown in ESM Tables 2–4. SNPs related to cardiometabolic diseases and the associations with MDD are shown in ESM Tables 5–7.
Statistical analyses
We calculated a Wald ratio estimate for each genetic variant and summarised the estimates using the random-effects inverse-variance weighted (IVW) method. The IVW method with random effects provides a concise estimation and takes into account potential heterogeneity among the Wald ratio estimates from individual SNPs [18]. Furthermore, we conducted several sensitivity analyses by using the weighted median, MR-Egger regression and MR pleiotropy residual sum and outlier (MR-PRESSO) methods as well as leave-one-out analysis. ORs and CIs were scaled to one-unit increment of loge OR of MDD, type 2 diabetes, CAD and heart failure.
The power calculation was based on the results of an online tool using several parameters, including sample size of the outcome GWAS, variance explained by selected SNPs and expected effect size [19]. The ‘get-or-from-lor’ commend in the TwoSampleMR package was used to estimate the explained variance for binary traits. The SNPs that were unavailable in the outcome datasets were replaced by proxies at R2 > 0.90. SNPs with a minor allele frequency above 0.42 were considered to be strand-ambiguous and removed from the analysis. The Bonferroni method was used to correct for multiple testing and, therefore, we considered associations with p values below 0.0083 (0.05/6) as strong evidence of associations. Results with p values between 0.0083 and 0.05 were regarded as suggestive associations. All analyses were two-sided and conducted using the TwoSampleMR package (https://mrcieu.github.io/TwoSampleMR/) in R software (version 3.6.0; www.r-project.org/).
Results
Causal effect of MDD on cardiometabolic diseases
Among the 96 MDD-associated SNPs, six were not available in the dataset of type 2 diabetes, and five of the six unavailable SNPs were replaced by suitable proxy SNPs. Furthermore, six SNPs were excluded due to ambiguous palindrome, resulting in 89 SNPs as instrumental variables in the analysis of type 2 diabetes and 90 SNPs in the analyses of CAD and heart failure (ESM Tables 2–4). The statistical power to detect an OR of 1.2 was 80% in the analysis of type 2 diabetes but only 55% and 63%, respectively, in the analyses of CAD and heart failure (ESM Table 8).
Genetic liability to MDD was significantly associated with type 2 diabetes and CAD risk and was suggestively associated with heart failure risk (Fig. 2). The OR of type 2 diabetes per one-unit increase in loge odds of MDD liability was 1.26 (95% CI 1.10, 1.43; p = 6 × 10−4) in the random-effects IVW model. The association was consistent across sensitivity analyses, albeit non-significant in the MR-Egger regression analysis. The MR-PRESSO analysis detected four outliers and provided an OR of type 2 diabetes of 1.22 (95% CI 1.09, 1.37; p = 8 × 10−4) after outlier correction. The Cochran’s Q value implied a substantial heterogeneity among estimates obtained from individual SNPs (Q = 333, p < 0.001) but the MR-Egger regression analysis yielded no indication of potential non-balanced pleiotropy (CI for intercept, −0.013, 0.013; p = 0.99). Moreover, leave-one-out analysis indicated that the observed association was not driven by any single SNP (ESM Fig. 1). The association was slightly weaker and borderline significant in a sensitivity analysis using SNPs associated with type 2 diabetes in the BMI-adjusted model (OR 1.15 [95% CI 1.03, 1.30]; p = 0.0166) (ESM Fig. 2).
The ORs per one-unit increase in loge odds of MDD liability were 1.16 (95% CI 1.05, 1.29; p = 0.005) for CAD and 1.11 (95% CI 1.01, 1.21; p = 0.033) for heart failure (Fig. 2). There was no observed heterogeneity (p of Cochran’s Q > 0.05), no pleiotropy (p for intercept, >0.05) and no outliers in the analysis of CAD or heart failure. Leave-one-out analysis indicated that the association with CAD was not driven by any single SNP (ESM Fig. 3) but six outlier SNPs were identified in the analysis of heart failure (ESM Fig. 4). We proceeded and searched for traits associated with those six outliers in a GWAS catalogue but found no likely trait that could confound the observed association between MDD and heart failure (ESM Table 4).
Causal effect of cardiometabolic diseases on MDD
After palindromic SNP exclusion and proxy replacement, we used 184 SNPs for type 2 diabetes, 37 SNPs for CAD and ten SNPs for heart failure as instrumental variables in the analysis of the effects of cardiometabolic diseases on MDD (ESM Tables 5–7). We had high power (over 90% power to detect an OR of 1.1) to detect weak associations of type 2 diabetes and CAD with MDD, but lower power in the analysis of the effect of heart failure on MDD (87% power to detect an OR of 1.20) (ESM Table 8). None of the three cardiometabolic diseases were associated with MDD, with ORs close to 1 (Fig. 3). Although there was suggestive evidence of an association between type 2 diabetes and MDD in the MR-Egger analysis (OR 0.93 [95% CI 0.88, 1.00]; p = 0.040), no association was observed after correction for three outliers (OR 0.99 [95% CI 0.97, 1.02]; p = 0.700) in the MR-PRESSO analysis (Fig. 3). Furthermore, there was no association between type 2 diabetes and MDD in sensitivity analyses using all 403 SNPs or using the 172 SNPs from the BMI-adjusted model (ESM Fig. 5).
Discussion
In the present bidirectional two-sample MR study, we observed a significant positive association of genetic liability to MDD with type 2 diabetes and CAD, and a suggestive association with heart failure. The reverse MR analysis provided no evidence that liability to type 2 diabetes, CAD or heart failure was related to MDD.
Adverse effects of MDD on type 2 diabetes development have been demonstrated consistently in observational studies. A meta-analysis of nine prospective studies proposed that depression was a risk factor for the onset of type 2 diabetes and reported a pooled RR of 1.26 (95% CI 1.13, 1.39), which is in agreement with our findings [20]. Results of another updated meta-analysis, which included 13 prospective studies with 6916 incident cases of type 2 diabetes, showed that having depression increased the risk of type 2 diabetes by 60% [21]. Additionally, a positive association between MDD and type 2 diabetes was supported by three large-scale cohort studies with medium or long follow-up periods (individual studies included 65,381 women followed up for 10 years, 11,694 adults for 6 years, and 5201 adults for 3.2 years) [22,23,24]. In contrast, epidemiological data on the effect of type 2 diabetes on MDD risk are inconclusive [25]. A systematic review including 83 studies found that type 2 diabetes could not independently predict MDD risk [26], but some recently published studies reported a significant positive association in both directions [21,22,23,24]. The present MR study found no evidence supporting a causal detrimental effect of type 2 diabetes on MDD. A cohort study that followed 65,381 women for 10 years found that the risk of developing clinical depression was higher in diabetic women, particularly in those who received insulin therapy, compared with non-diabetic women [22]. Another cohort study reported that treated but not untreated type 2 diabetes was associated with an increased risk of depression [23]. These studies suggest that MDD risk in type 2 diabetes may differ depending on type 2 diabetes treatment [22, 23].
We found evidence that MDD may be causally associated with risk of CAD and possibly heart failure, thereby confirming the results of observational studies [9,10,11,12, 27]. Notably, diabetes and CAD have been proposed as risk factors for heart failure, and CAD can explain more than 60% of heart failure cases [28]. Thus, the suggestive association between MDD and heart failure might be partly mediated via type 2 diabetes and CAD.
There are several possible mechanisms that can explain the causal effect of MDD on type 2 diabetes risk. A series of biological abnormalities related to depression, including increased counter-regulatory hormone release and activity, alterations in glucose transport function and increased immunoinflammatory activation, may influence the risk of type 2 diabetes [5]. In addition, lifestyle factors, such as smoking and alcohol consumption, may play a mediating role in the pathway from depression to type 2 diabetes [29].
In traditional observational studies of the effect of depression on type 2 diabetes, the results are prone to be biased by reverse causality, as impaired glucose tolerance may lead to depression before diabetes symptoms manifest. Antidepressant medication, which has been demonstrated to be associated with hyperglycaemia, may introduce residual confounding [25]. We used the MR study design, thereby avoiding reverse causality and minimising residual confounding. Another strength is that we extracted summary-level data from the hitherto largest GWAS for MDD and type 2 diabetes and, therefore, we had high power to detect even weak associations. A limitation of the present study is the potential of pleiotropy. However, we detected no directional pleiotropy in the MR-Egger regression analysis and the estimates were consistent when using the weighted median and MR-PRESSO analyses, which indicated a negligible distortion by potential pleiotropy. A previous MR study revealed that BMI was a risk factor for both MDD [30] and type 2 diabetes [31], indicating that BMI might be a pleiotropic factor or confounder in the pathway from MDD to type 2 diabetes. However, in a sensitivity analysis using BMI-adjusted estimates for the genetic associations with type 2 diabetes, the effect of MDD on type 2 diabetes remained at the conventional level of significance (p < 0.05). This implies that the causal association between MDD and type 2 diabetes was not completely mediated or biased by BMI. Another limitation is that population stratification may have affected the result for the association between CAD and MDD because the GWAS of CAD included participants of different ancestry. However, as the majority (77%) of individuals were of European ancestry, any population stratification bias was expected to be small.
The present MR study strengthens the evidence that MDD is a potential risk factor for type 2 diabetes and CAD. However, there was no genetic support for a causal effect of type 2 diabetes or CAD on MDD. Whether MDD is causally related to heart failure needs further investigation. Given the high disease burden related to the causal link, it is recommended that MDD prevention, management and treatment should be enhanced for type 2 diabetes prevention.
Data availability
All data included in this study were uploaded as supplements and are also publicly available through published GWASs and open GWAS datasets (UK Biobank, 23andMe and Psychiatric Genomics: https://datashare.is.ed.ac.uk/handle/10283/3203; DIAGRAM: http://diagram-consortium.org/downloads.html; CARDIoGRAMplusCD4: www.cardiogramplusc4d.org/; HERMES: http://www.kp4cd.org/datasets/mi).
Abbreviations
- CAD:
-
Coronary artery disease
- GWAS:
-
Genome-wide association study
- IVW:
-
Inverse-variance weighted
- MDD:
-
Major depressive disorder
- MR:
-
Mendelian randomisation
- MR-PRESSO:
-
Mendelian randomisation pleiotropy residual sum and outlier
References
World Health Organization (2017) Depression and other common mental disorders: global health estimates. World Health Organization, Geneva
Ogurtsova K, da Rocha Fernandes JD, Huang Y et al (2017) IDF diabetes atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract 28:40–50
Anderson RJ, Freedland KE, Clouse RE, Lustman PJ (2001) The prevalence of comorbid depression in adults with diabetes. Diabetes Care 24(6):1069–1078. https://doi.org/10.2337/diacare.24.6.1069
Carnethon MR, Kinder LS, Fair JM, Stafford RS, Fortmann SP (2003) Symptoms of depression as a risk factor for incident diabetes: findings from the National Health and Nutrition Examination Epidemiologic Follow-up Study, 1971–1992. Am J Epidemiol 158(5):416–423. https://doi.org/10.1093/aje/kwg172
Musselman DL, Betan E, Larsen H, Phillips LS (2003) Relationship of depression to diabetes types 1 and 2: epidemiology, biology, and treatment. Biol Psychiatry 54(3):317–329. https://doi.org/10.1016/s0006-3223(03)00569-9
de Jonge P, Roy JF, Saz P, Marcos G, Lobo A, ZARADEMP Investigators (2006) Prevalent and incident depression in community-dwelling elderly persons with diabetes mellitus: results from the ZARADEMP project. Diabetologia 49(11):2627–2633. https://doi.org/10.1007/s00125-006-0442-x
Tabák AG, Akbaraly TN, Batty GD, Kivimäki M (2014) Depression and type 2 diabetes: a causal association? Lancet Diabetes Endocrinol 2(3):236–245. https://doi.org/10.1016/S2213-8587(13)70139-6
Smith G, Ebrahim S (2003) ʻMendelian randomizationʼ: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32(1):1–22. https://doi.org/10.1093/ije/dyg070
Abramson J, Berger A, Krumholz HM, Vaccarino V (2001) Depression and risk of heart failure among older persons with isolated systolic hypertension. JAMA Intern Med 161(14):1725–1730
Lett HS, Blumenthal JA, Babyak MA et al (2004) Depression as a risk factor for coronary artery disease: evidence, mechanisms, and treatment. Psychosom Med 66(3):305–315. https://doi.org/10.1097/01.psy.0000126207.43307.c0
Rugulies R (2002) Depression as a predictor for coronary heart disease: a review and meta-analysis. Am J Prev Med 23(1):51–61. https://doi.org/10.1016/S0749-3797(02)00439-7
Williams SA, Kasl SV, Heiat A, Abramson JL, Krumholz HM, Vaccarino V (2002) Depression and risk of heart failure among the elderly: a prospective community-based study. Psychosom Med 64(1):6–12. https://doi.org/10.1097/00006842-200201000-00002
Davey Smith G, Ebrahim S (2005) What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ 330(7499):1076–1079. https://doi.org/10.1136/bmj.330.7499.1076
Howard DM, Adams MJ, Clarke T-K et al (2019) Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22(3):343–352. https://doi.org/10.1038/s41593-018-0326-7
Mahajan A, Taliun D, Thurner M et al (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50(11):1505–1513. https://doi.org/10.1038/s41588-018-0241-6
Nikpay M, Goel A, Won H-H et al (2015) A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 47(10):1121–1130. https://doi.org/10.1038/ng.3396
Shah S, Henry A, Roselli C et al (2020) Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 11(1):163. https://doi.org/10.1038/s41467-019-13690-5
Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan N, Thompson J (2017) A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 36(11):1783–1802. https://doi.org/10.1002/sim.7221
Brion M-JA, Shakhbazov K, Visscher PM (2012) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42(5):1497–1501
Knol MJ, Twisk JWR, Beekman ATF, Heine RJ, Snoek FJ, Pouwer F (2006) Depression as a risk factor for the onset of type 2 diabetes mellitus. A meta-analysis. Diabetologia 49(5):837–845. https://doi.org/10.1007/s00125-006-0159-x
Mezuk B, Eaton WW, Albrecht S, Golden SH (2008) Depression and type 2 diabetes over the lifespan. A meta-analysis. 31(12):2383–2390
Pan A, Lucas M, Sun Q et al (2010) Bidirectional association between depression and type 2 diabetes mellitus in women. JAMA Intern Med 170(21):1884–1891
Golden SH, Lazo M, Carnethon M et al (2008) Examining a bidirectional association between depressive symptoms and diabetes. JAMA 299(23):2751–2759. https://doi.org/10.1001/jama.299.23.2751
Chen P-C, Chan Y-T, Chen H-F, Ko M-C, Li C-Y (2013) Population-based cohort analyses of the bidirectional relationship between type 2 diabetes and depression. Diabetes Care 36(2):376–382. https://doi.org/10.2337/dc12-0473
Renn BN, Feliciano L, Segal DL (2011) The bidirectional relationship of depression and diabetes: a systematic review. Clin Psychol Rev 31(8):1239–1246. https://doi.org/10.1016/j.cpr.2011.08.001
Talbot F, Nouwen A (2000) A review of the relationship between depression and diabetes in adults: is there a link? Diabetes Care 23(10):1556–1562. https://doi.org/10.2337/diacare.23.10.1556
Khawaja IS, Westermeyer JJ, Gajwani P, Feinstein RE (2009) Depression and coronary artery disease: the association, mechanisms, and therapeutic implications. Psychiatry 6(1):38–51
He J, Ogden LG, Bazzano LA, Vupputuri S, Loria C, Whelton PK (2001) Risk factors for congestive heart failure in US men and women: NHANES I Epidemiologic Follow-up Study. JAMA Intern Med 161(7):996–1002
Strine TW, Mokdad AH, Dube SR et al (2008) The association of depression and anxiety with obesity and unhealthy behaviors among community-dwelling US adults. Gen Hosp Psychiatry 30(2):127–137. https://doi.org/10.1016/j.genhosppsych.2007.12.008
Tyrrell J, Mulugeta A, Wood AR et al (2019) Using genetics to understand the causal influence of higher BMI on depression. Int J Epidemiol 48(3):834–848. https://doi.org/10.1093/ije/dyy223
Wainberg M, Mahajan A, Kundaje A et al (2019) Homogeneity in the association of body mass index with type 2 diabetes across the UK Biobank: a Mendelian randomization study. PLoS Med 16(12):e1002982. https://doi.org/10.1371/journal.pmed.1002982
Acknowledgements
Summary-level data for genetic associations with MDD, type 2 diabetes, CAD and heart failure were from the MDD GWAS dataset (the UK Biobank study, 23andMe and Psychiatric Genomics consortium), the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium, the Coronary ARtery DIsease Genome wide Replication and Meta-analysis plus The Coronary Artery Disease Genetics (CARDIoGRAMplusC4D) consortium and the Heart Failure Molecular Epidemiology for Therapeutic Targets (HERMES) consortium. The authors thank all investigators for sharing these data.
Authors’ relationships and activities
The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Funding
Open access funding provided by Uppsala University. This work was supported by the Swedish Heart-Lung Foundation (Hjärt-Lungfonden grant no. 20190247) and the Swedish Research Council (Vetenskapsrådet grant no. 2019–00977).
Author information
Authors and Affiliations
Contributions
BT, SY and SCL designed the study. BT, SY and YX analysed the data and wrote the manuscript. BT, SY, SCL, YX and QH interpreted the data and revised the paper. All authors have given final approval of the version to be published. SCL is the guarantor of this study.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1770 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tang, B., Yuan, S., Xiong, Y. et al. Major depressive disorder and cardiometabolic diseases: a bidirectional Mendelian randomisation study. Diabetologia 63, 1305–1311 (2020). https://doi.org/10.1007/s00125-020-05131-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-020-05131-6