Abstract
Key message
Twenty-eight QTLs for LLS disease resistance were identified using an amphidiploid constructed mapping population, a favorable 530-kb chromosome segment derived from wild species contributes to the LLS resistance.
Abstract
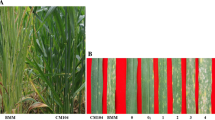
Late leaf spot (LLS) is one of the major foliar diseases of peanut, causing serious yield loss and affecting the quality of kernel and forage. Some wild Arachis species possess higher resistance to LLS as compared with cultivated peanut; however, ploidy level differences restrict utilization of wild species. In this study, a synthetic amphidiploid (Ipadur) of wild peanuts with high LLS resistance was used to cross with Tifrunner to construct TI population. In total, 200 recombinant inbred lines were collected for whole-genome resequencing. A high-density bin-based genetic linkage map was constructed, which includes 4,809 bin markers with an average inter-bin distance of 0.43 cM. The recombination across cultivated and wild species was unevenly distributed, providing a novel recombination landscape for cultivated-wild Arachis species. Using phenotyping data collected across three environments, 28 QTLs for LLS disease resistance were identified, explaining 4.35–20.42% of phenotypic variation. The major QTL located on chromosome 14, qLLS14.1, could be consistently detected in 2021 Jiyang and 2022 Henan with 20.42% and 12.12% PVE, respectively. A favorable 530-kb chromosome segment derived from Ipadur was identified in the region of qLLS14.1, in which 23 disease resistance proteins were located and six of them showed significant sequence variations between Tifrunner and Ipadur. Allelic variation analysis indicating the 530-kb segment of wild species might contribute to the disease resistance of LLS. These associate genomic regions and candidate resistance genes are of great significance for peanut breeding programs for bringing durable resistance through pyramiding such multiple LLS resistance loci into peanut cultivars.




Similar content being viewed by others
Data availability
All information is specified in the manuscript or included as Additional Files.
References
Agarwal G, Clevenger J, Pandey MK, Wang H, Shasidhar Y, Chu Y, Fountain JC, Choudhary D, Culbreath AK, Liu X, Huang G, Wang X, Deshmukh R, Holbrook CC, Bertioli DJ, Ozias-Akins P, Jackson SA, Varshney RK, Guo B (2018) High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut. Plant Biotechnol J 16:1954–1967
Ahmad S, Nawade B, Sangh C, Mishra GP, Bosamia TC, Kumar N, Dobaria JR, Gajera HP (2020) Identification of novel QTLs for late leaf spot resistance and validation of a major rust QTL in peanut (Arachis hypogaea L.). 3 Biotech. 10:458
Chaudhari S, Khare D, Patil SC, Sundravadana S, Variath MT, Sudini HK, Manohar SS, Bhat RS, Pasupuleti J (2019) Genotype x environment studies on resistance to late leaf spot and rust in genomic selection training population of peanut (Arachis hypogaea L.). Front Plant Sci 10:1338
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, Li Y, Ye J, Yu C, Li Z, Zhang X, Wang J, Yang H, Fang L, Chen Q (2018) SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7:1–6
Chu Y, Chee P, Culbreath A, Isleib TG, Holbrook CC, Ozias-Akins P (2019) Major QTLs for resistance to early and late leaf spot diseases are identified on chromosomes 3 and 5 in peanut (Arachis hypogaea). Front Plant Sci 10:883
Clevenger J, Chu Y, Chavarro C, Botton S, Culbreath A, Isleib TG, Holbrook CC, Ozias-Akins P (2018) Mapping late leaf spot resistance in peanut (Arachis hypogaea) Using QTL-seq reveals markers for marker-assisted selection. Front Plant Sci 9:83
Favero AP, Dos Santos RF, Simpson CE, Valls JF, Vello NA (2015) Successful crosses between fungal-resistant wild species of Arachis (section Arachis) and Arachis hypogaea. Genet Mol Biol 38:353–365
Gangurde SS, Nayak SN, Joshi P, Purohit S, Sudini HK, Chitikineni A, Hong Y, Guo B, Chen X, Pandey MK, Varshney RK (2021) Comparative transcriptome analysis identified candidate genes for late leaf spot resistance and cause of defoliation in groundnut. Int J Mol Sci 22(9):4491
Gonda I, Ashrafi H, Lyon DA, Strickler SR, Hulse-Kemp AM, Ma Q, Sun H, Stoffel K, Powell AF, Futrell S, Thannhauser TW, Fei Z, Van Deynze AE, Mueller LA, Giovannoni JJ, Foolad MR (2019) Sequencing-based bin map construction of a tomato mapping population, facilitating high-resolution quantitative trait loci detection. Plant Genom 12(1):180010
Han S, Yuan M, Clevenger JP, Li C, Hagan A, Zhang X, Chen C, He G (2018) A SNP-based linkage map revealed QTLs for resistance to early and late leaf spot diseases in peanut (Arachis hypogaea L.). Front Plant Sci 9:1012
He Q, Zhi H, Tang S, Xing L, Wang S, Wang H, Zhang A, Li Y, Gao M, Zhang H, Chen G, Dai S, Li J, Yang J, Liu H, Zhang W, Jia Y, Li S, Liu J, Qiao Z, Guo E, Jia G, Liu J, Diao X (2021) QTL mapping for foxtail millet plant height in multi-environment using an ultra-high density bin map. Theor Appl Genet 134:557–572
Holbrook CC, Anderson WF (1995) Evaluation of a core collection to identify resistance to late leaf spot in peanut. Crop Sci 35:1700–1702
Jiang Y, Luo H, Yu B, Ding Y, Kang Y, Huang L, Zhou X, Liu N, Chen W, Guo J, Huai D, Lei Y, Jiang H, Yan L, Liao B (2021) High-density genetic linkage map construction using whole-genome resequencing for mapping QTLs of resistance to Aspergillus flavus infection in peanut. Front Plant Sci 12:745408
Khera P, Pandey MK, Wang H, Feng S, Qiao L, Culbreath AK, Kale S, Wang J, Holbrook CC, Zhuang W, Varshney RK, Guo B (2016) Mapping quantitative trait loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022. PLoS ONE 11:e0158452
Kolekar RM, Sujay V, Shirasawa K, Sukruth M, Khedikar YP, Gowda MVC, Pandey MK, Varshney RK, Bhat RS (2016) QTL mapping for late leaf spot and rust resistance using an improved genetic map and extensive phenotypic data on a recombinant inbred line population in peanut (Arachis hypogaea L.). Euphytica 209:147–156
Kumar KR, Kirti PB (2011) Differential gene expression in Arachis diogoi upon interaction with peanut late leaf spot pathogen, Phaeoisariopsis personata and characterization of a pathogen induced cyclophilin. Plant Mol Biol 75:497–513
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinform 25:1754–1760
Li W, Huang L, Liu N, Chen Y, Guo J, Yu B, Luo H, Zhou X, Huai D, Chen W, Yan L, Wang X, Lei Y, Liao B, Jiang H (2023) Identification of a stable major sucrose-related QTL and diagnostic marker for flavor improvement in peanut. Theor Appl Genet 136:78
Li W, Liu N, Huang L, Chen Y, Guo J, Yu B, Luo H, Zhou X, Huai D, Chen W, Yan L, Wang X, Lei Y, Liao B, Jiang H (2022) Stable major QTL on chromosomes A07 and A08 increase shelling percentage in peanut (Arachis hypogaea L.). The Crop J 10:820–829
Liang Y, Baring M, Wang S, Septiningsih EM (2017) Mapping QTLs for leafspot resistance in peanut using SNP-based next-generation sequencing markers. Plant Breed Biotechnol 5:115–122
Mehan VK, Reddy PM, Subrahmanyam P, McDonald D, Singh AK (1996) Identification of new sources of resistance to rust and late leaf spot in peanut∗. Int J Pest Manag 42:267–271
Pandey MK, Khan AW, Singh VK, Vishwakarma MK, Shasidhar Y, Kumar V, Garg V, Bhat RS, Chitikineni A, Janila P, Guo B, Varshney RK (2017a) QTL-seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnol J 15:927–941
Pandey MK, Monyo E, Ozias-Akins P, Liang X, Guimaraes P, Nigam SN, Upadhyaya HD, Janila P, Zhang X, Guo B, Cook DR, Bertioli DJ, Michelmore R, Varshney RK (2012) Advances in Arachis genomics for peanut improvement. Biotechnol Adv 30:639–651
Pandey MK, Wang H, Khera P, Vishwakarma MK, Kale SM, Culbreath AK, Holbrook CC, Wang X, Varshney RK, Guo B (2017b) Genetic dissection of novel QTLs for resistance to leaf spots and tomato spotted wilt virus in peanut (Arachis hypogaea L.). Front Plant Sci 8:25
Shirasawa K, Bhat RS, Khedikar YP, Sujay V, Kolekar RM, Yeri SB, Sukruth M, Cholin S, Asha B, Pandey MK, Varshney RK, Gowda MVC (2018) Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L.). Front Plant Sci 9:1727
Singh MP, Erickson JE, Boote KJ, Tillman BL, Jones JW, Bruggen AHC (2011) Late leaf spot effects on growth, photosynthesis, and yield in peanut cultivars of differing resistance. Agron J 103:85–91
Stalker HT (2017) Utilizing wild species for peanut improvement. Crop Sci 57:1102–1120
Stalker HT, Tallury SP, Ozias-Akins P, Bertioli D, Bertioli SCL (2013) The value of diploid peanut relatives for breeding and genomics. Peanut Sci 40:70–88
Subrahmanyam P, Rao VR, McDonald D, Moss JP, Gibbons RW (1989) Origins of resistances to rust and late leaf spot in peanut (Arachis Hypogaea, Fabaceae). Econ Bot 43:444–455
Tallury SP, Isleib TG, Copeland SC, Rosas-Anderson P, Balota M, Singh D, Stalker HT (2014) Registration of two multiple disease-resistant peanut germplasm lines derived from Arachis cardenasii Krapov. & W.C. Gregory, GKP 10017. J Plant Regist 8:86–89
Wang H, Pandey MK, Qiao L, Qin H, Culbreath AK, He G, Varshney RK, Scully BT, Guo B (2013) Genetic mapping and quantitative trait loci analysis for disease resistance using F2 and F5 generation-based genetic maps derived from ‘Tifrunner’ × ‘GT-C20’ in peanut. Plant Genom. https://doi.org/10.3835/plantgenome2013.05.0018
Wu Y, Bhat PR, Close TJ, Lonardi S (2008) Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4:e1000212
Zhou X, Xia Y, Liao J, Liu K, Li Q, Dong Y, Ren X, Chen Y, Huang L, Liao B, Lei Y, Yan L, Jiang H (2016) Quantitative trait locus analysis of late leaf spot resistance and plant-type-related traits in cultivated peanut (Arachis hypogaea L.) under multi-environments. PLoS ONE 11:e0166873
Acknowledgements
This research is supported by National Natural Science Foundation of China (31861143009), Guangxi Science and Technology Major Project (“Peak Project” of Modern Characteristic Agriculture, GuikeAA23062004), National Key Research and Development Program of China (2022YFD1200400), Key Research and Development Project of Shandong Province (2020LZGC001, 2022LZGC007), Agricultural scientific and technological innovation project of SAAS (CXGC2023C04, CXGC2023G30), and Taishan Scholar Project of Shandong Province
Funding
This study was funded by the National Natural Science Foundation of China (31861143009), Guangxi Science and Technology Major Project (“Peak Project” of Modern Characteristic Agriculture, GuikeAA23062004), National Key Research and Development Program of China (2022YFD1200400, 2023YFD1202800), Key Research and Development Project of Shandong Province (2020LZGC001, 2022LZGC007), New 20 Policies for Universities in Jinan (202333047), Agricultural Scientific and Technological Innovation Project of SAAS (CXGC2023C04, CXGC2023G30), and Taishan Scholar Project of Shandong Province.
Author information
Authors and Affiliations
Contributions
CZ and XW conceived and designed the experiments. JP, XL, CF, ZW, CY, RT, XS and MG performed the experiments. CL, HX, SZ, LH, HZ, DB and SL developed the populations. JB, XL and GW provided technical assistance and some analysis. JP and CZ wrote the manuscript. MP revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest, and the manuscript is approved by all authors for publication.
Additional information
Communicated by Reyazul Rouf Mir.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2024_4580_MOESM2_ESM.pdf
Recombination bin map of 200 individual RILs. Colored tracks represent the 200 RILs of the TI population that were used for bin map construction: blue: genotype inherited from maternal parent Tifrunner, red: genotype inherited from maternal parent Ipadur, yello: genotype inherited from heterozygous genotype (Tifrunner × Ipadur) F1.
122_2024_4580_MOESM3_ESM.pdf
Collinearity analysis between genetic map and physical map. The x-axis scales the physical positions of markers based on reference sequences. The y-axis represents the genetic distance of the markers in centimorgans accordingly.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, J., Li, X., Fu, C. et al. High-density bin-based genetic map reveals a 530-kb chromosome segment derived from wild peanut contributing to late leaf spot resistance. Theor Appl Genet 137, 69 (2024). https://doi.org/10.1007/s00122-024-04580-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04580-6