Abstract
Key message
Two major quantitative trait loci (QTLs) and five minor QTLs for 10 pathotypes were identified on chromosomes C01, C03, C04 and C08 through genotyping-by-sequencing from Brassica oleracea.
Abstract
Clubroot caused by Plasmodiophora brassicae is an important disease in brassica crops. Managing clubroot disease of canola on the Canadian prairie is challenging due to the continuous emergence of new pathotypes. Brassica oleracea is considered a major source of quantitative resistance to clubroot. Genotyping-by-sequencing (GBS) was performed in the parental lines; T010000DH3 (susceptible), ECD11 (resistant) and 124 BC1 plants. A total of 4769 high-quality polymorphic SNP loci were obtained and distributed on 9 chromosomes of B. oleracea. Evaluation of 124 BC1S1 lines for resistance to 10 pathotypes: 3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O of P. brassicae, was carried out. Seven QTLs, 5 originating from ECD11 and 2 from T010000DH3, were detected. One major QTL designated as Rcr_C03-1 on C03 contributed 16.0–65.6% of phenotypic variation explained (PVE) for 8 pathotypes: 2B, 5C, 5G, 3H, 8J, 5K, 5L and 3O. Another major QTL designated as Rcr_C08-1 on C08 contributed 8.3 and 23.5% PVE for resistance to 8J and 5K, respectively. Five minor QTLs designated as Rcr_C01-1, Rcr_C03-2, Rcr_C03-3, Rcr_C04-1 and Rcr_C08-2 were detected on chromosomes C01, C03, C04 and C08 that contributed 8.3–23.5% PVE for 5 pathotypes each of 3A, 2B, 3D, 8J and 5K. There were 1, 10 and 4 genes encoding TIR-NBS-LRR/CC-NBS-LRR class disease resistance proteins in the Rcr_C01-1, Rcr_C03-1 and Rcr_C08-1 flanking regions. The syntenic regions of the two major QTLs Rcr_C03-1 and Rcr_C08-1 in the B. rapa genome ‘Chiifu’ were searched.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica species are grown as source of oil, vegetable, condiment, and fodder worldwide. The ‘Triangle of U’ (Morinaga 1934; Nagaharu and Nagaharu 1935) revealed relationships among the main species of Brassica genus, which makes more interesting of these species to the researchers for genomic utilization among the species. B. napus (AACC; n = 19), B. juncea (AABB; n = 18) and B. carinata (BBCC; n = 17) are amphidiploid species resulting from hybridization between pairs of the diploid species: B. rapa (AA; n = 10), B. nigra (BB; n = 8) and B. oleracea (CC; n = 9).
Clubroot, caused by Plasmodiophora brassicae Woronin, a soil-borne obligate biotrophic protist, is an important disease in brassica crops worldwide. The pathogen causes club or spindle shape gall formation on roots, which hampers nutrients and water uptake for plants growth. The disease has been reported in more than 60 countries, causing about 10–15% yield loss globally (Dixon 2009). The disease not only caused yield loss but also affected the quality of the crops (Engqvist 1994; Pageau et al. 2006). Some practices such as liming and early spring seeding can reduce clubroot levels but is not economical for large-scale canola production (Voorrips et al. 1995; Gossen et al. 2012, Donald et al. 2014; Hwang et al. 2014). Utilization of clubroot resistant (CR) cultivars is the most effective way to control the disease (Peng et al. 2014; Rahman et al. 2014). However, repeated utilization of the same CR source is not a good strategy as it provides opportunity to minor pathotypes to become predominant pathotypes (Sedaghatkish et al. 2019; Cao et al. 2020). In Canada, breeding companies released CR varieties in 2009–2010 after identifying clubroot disease in canola fields in 2003 (Tewari et al. 2005). Most CR canola cultivars probably were developed by using a CR gene from the oilseed rape ‘Mendel’ (Fredua-Agyeman et al. 2018), which became susceptible just after 4 years in 2013 due to emergence of new virulent pathotype 5X (Strelkov et al. 2016). This incidence led to closely focus on pathogen populations in Canadian canola fields. As a result, > 30 pathotypes have been classified based on the Canadian Clubroot Differential (CCD) set (Strelkov et al. 2018; Hollman et al. 2021). In addition to the most prevalent original pathotype 3H, current predominant new pathotypes 3A, 3D (Dakouri et al. 2021) and other pathotypes are also needed to consider to find new resistant sources. Along with crop rotation, gene pyramiding from different sources is considered an effective strategy for successful control of clubroot disease caused by different pathotypes of the pathogen.
As resistance sources to clubroot in B. napus (AACC) are very limited, researchers have searched resistance in its diploid progenitor species B. rapa (AA) and B. oleracea (CC). Genes can be transferred from B. rapa (AA) and B. oleracea (CC) to B. napus either interspecific breeding or developing new re-synthesized B. napus. CR in B. rapa was usually controlled by major genes, while CR in B. oleracea was mostly reported as quantitative trait loci (QTL), polygenic control. Although polygenic resistance is considered more durable and can be race non-specific (Diederichsen et al. 2009), resistance sources in B. oleracea are very limited (Dias et al. 1993; Manzanares-Dauleux et al. 2000; Voorrips 1995; Diederichsen et al. 2009). Cabbage (B. oleracea var. capitata) cultivar ‘Badger Shipper’ (ECD11) showed resistance to 15 pathotypes out of 17 pathotypes in the CCD Set (Strelkov et al. 2018), which can be very useful as race non-specific CR source.
Identification of QTLs for resistance to clubroot has been performed in B. oleracea. Four QTLs, qCRc4-1 on C04, qCRc7-2, qCRc7-3, and qCRc7-4 on C07 were identified through QTL-Seq for resistance to race 4 of P. brassicae in cabbage breeding line GZ87 (Ce et al. 2021). Another two QTLs, DIC.I-1 and DIC.II-1, were identified on C08 from the same B. oleracea inbred line GZ87 against race 4 (Peng et al. 2018). Ten QTLs; PbC4.1, PbC6, PbC7.1, PbC7.2, PbC8, PbC9.1, PbC3, PbC4.2, PbC7.3 and PbC9.2 from six C-genome chromosomes; C03, C04, C06, C07, C08 and C09 were identified through association mapping against pathotype 3A and 5X-LG2 (Farid et al. 2020). Two QTLs, CRQTL-GN_1 and CRQTL-GN_2 on C02 and C03 for resistance to race 9, and one QTL, CRQTL-YC on C03 for resistance race 2 were identified (Lee et al. 2016). One major QTL, pb-Bo(Anju)1 on C02, four minor QTLs pb-Bo(Anju)2 on C02, pb-Bo(Anju)3 on C03, pb-Bo(Anju)4 on C07 and pb-Bo(GC)1 on C05 were derived from resistant line Anju and susceptible line GC (Nagaoka et al. 2010). A total of nine QTLs, Pb-Bo1, Pb-Bo2, Pb-Bo3, Pb-Bo4, Pb-Bo5a, Pb-Bo5b, Pb-Bo8, Pb-Bo9a and Pb-Bo9b, were identified from a selected line (C10) of a French kale landrace (B. oleracea var. acephala) for clubroot resistance on C01–05, C08–09 against five isolates, Ms6 and eH (pathotype P1), K92 (pathotype P2), K92-16 (pathotype P4), Pb137-522 (pathotype P7) of P. brassicae. Two to five QTLs were detected depending on the isolates. The Pb-Bo1 was detected in all isolates and explained 20.7–80.7% of the phenotypic variation (Rocherieux et al. 2004). A QTL was identified on C03 from a inbreed kale line K269 against pathotype 1 and 3 (Moriguchi et al. 1999). Two QTLs, pb-3 and pb-4, were identified from the German landrace ‘Bindsachsener’ on C03 and C01 against a field isolate characterized as ECD 16/3/30 (Voorrips et al. 1997). Only one major dominant CR gene (Rcr7) was identified from chromosome C7 originating from A-genome of B. rapa (Dakouri et al. 2018).
Although no CR gene has been cloned from C-genome, three CR genes CRa, Crr1 and CRbkato were cloned from A-genome, which encode toll-interleukin-1 receptor, nucleotide binding site and leucine-rich repeat (TIR-NBS-LRR) proteins (Ueno et al. 2012; Hatakeyama et al. 2013, 2017). Resistant genes encoded NBS-LRR protein families can be subdivided according to their functional domain as TIR domain containing (TNL) and coiled-coil (CC) domain containing (CNL) subfamilies (McHale et al. 2006). NBS-LRR-related disease resistance is effective against obligate and hemi-biotrophic pathogens (Glazebrook et al. 2005). Although TNL protein encoding genes are reported for CR resistance, no CR gene encoding CNL protein has been reported.
In this study, a BC1/BC1S1 mapping population was developed using resistant cabbage cultivar ECD11 crossed with clubroot-susceptible doubled haploid (DH) line T010000DH3. The objectives of the current study were: 1) to characterize the genome-wide DNA variants in the BC1 lines through genotyping-by-sequencing (GBS); 2) to identify QTLs associated with resistance to 10 pathotypes of P. brassicae characterized with the CCD set; and 3) to identify possible candidate genes for each QTL.
Materials and methods
Plant materials
Seeds of resistant cultivar ECD11 and susceptible DH line T010000DH3 derived from Chinese kale (B. oleracea) cultivar ‘TO1434’ were provided by Dr. G. R. Dixon (The University of Warwick, Wellesbourne, Warwick, UK) and Dr. I. Parkin (Saskatoon Research and Development Centre, Saskatoon, SK, Canada), respectively. ECD11 was crossed to T010000DH3 (pollen donor) to produce F1 progeny. Backcross with T010000DH3 (recurrent parent) was performed to produce the BC1 population. In total, 124 BC1 plants were used for GBS and self-pollinated to produce a BC1S1 population consisting of 124 lines for inoculation testing at the Saskatoon Research and Development Centre.
Evaluation of plants for resistance to clubroot
Strains of P. brassicae collected from canola fields in Alberta were provided by Dr. S.E. Strelkov at the University of Alberta, Canada. The inoculum preparation method used in the study was as described by Suwabe et al. (2003). Seedlings of a susceptible cultivar were inoculated and maintained under controlled conditions. Clubbed roots were harvested from infected plants after 5–6 weeks and stored at − 20 °C. Fresh inoculum of each pathotype was prepared after softening about 5 g of frozen club in a small amount of distilled water for 1–2 h. The material was homogenized in a blender for 2 min and strained through 2–3 layers of nylon mesh cloth. The resulting spore suspension was diluted with deionized water to produce a final concentration of 1 × 107 resting spores mL−1. Plants were tested for resistance to 10 pathotypes of P. brassicae (strain F.3–14 for pathotype 3A, F.183–14 for 2B, F.175–14 for 5C, F.1–14 for 3D, CDCS for 5G, P. 41–14 for 3H, F.12–15 for 8J, F.10–15 for 5K, CDCN#2 for 5L and F.381–16 for 3O). The inoculation method used in the current study was as described by Yu et al. (2021). Seeds of the BC1S1 population were sown into Sunshine #3 soilless mix (Sun Gro Horticulture Canada Ltd.; Seba Beach, AB) with Osmocote (Everris NA Inc.; Dublin, OH, USA) in 32-pots inserts held by trays (The HC Companies; Twinsburg, OH, USA). Adequate amount of water was added to each tray to soak the soilless mix overnight. Similar experimental design as described by Yu et al. (2017) was used in this study. Ten to twelve seeds were sown in each pot/line, which were inoculated with 15 ml of inoculum (1 × 107 spores/ml) after seven days of sowing. The tray holding pot was covered with a dome for two weeks. The inoculated plants were grown in a growth chamber set at 22/18 °C day/night temperature with a 16 h photoperiod. The pots were always kept moist by adding adequate amount of water whenever necessary. Leaves of the plants were pruned every alternate days after two weeks of seeding. Watering was stopped at 2 days before ratting clubroot. The B. rapa var. pekinensis ‘Granaat’ ECD05 (susceptible to all 10 pathotypes used in this study) and the parental lines (ECD11 and T010000DH3) were included as controls.
Six weeks after inoculation, each plant was rated for clubroot symptoms using a 0–3 scale (Kuginuki et al. 1999), where 0 = no symptoms, 1 = a few small clubs, 2 = moderate clubbing, and 3 = severe clubbing (Fig. S1). A disease severity index (DSI) was calculated for each line using the method of Horiuchi and Hori (1980):
Pearson correlation coefficient was calculated with disease severity indexes (DSIs) of 124 BC1S1 populations.
The F1 (T010000DH3 × ECD11) also included in the inoculation test for all the ten pathotypes in this study. Reciprocal F1 (ECD11 × T010000DH3) were tested with only 3 pathotypes 3A, 2B and 3D as only few seeds from the cross were obtained, so it was not possible to test all pathotypes. The experiment was repeated twice. Each of the repetitions provided a similar result in most cases. For those lines with inconsistent results, the higher DSI among the two repetitions of the assessment was considered to be more accurate and was used to characterize the resistance response of the line.
GBS of the parental lines and 124 BC1 plants
Young leaves of the 124 BC1 plants and two replications of parental lines were collected and freeze-dried in a Freezone 6 dryer (Labconco Corp, Kansas City, MO) for 48 h and grinded using the Mixture Mills 300 (Retsch Inc., Newtown, PA). DNA was extracted using DNeasy 96 Plant Kit (Qiagen, Toronto, ON, Canada) following DNeasy Plant Handbook from QIAGEN, quantified using a NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ), diluted to 10 ng/μl and kept at −20 °C until subsequent use for sequencing and genotyping. In total, 128 DNA samples were prepared for sequencing. GBS library generation, PstI/MspI for Illumina with size selection were done in Institute of integrative biology and systems (Université Laval, Québec, Canada). Illumina library QC and Illumina HiSeq2500 PE125 sequencing were performed at Genome Quebec (Montreal, Canada). Two replications of parental lines were performed to increase the sequencing depth, to provide a more accurate call of the genotype at each SNP site in the BC1 population.
Alignment of GBS short reads into B. oleracea reference genome sequence and SNP calling
The reference genome sequence of DH line ‘D134’ (cabbage; B. oleracea var. capitata) generated by third-generation sequencing technology (Lv et al. 2020) and downloaded from https://db.cngb.org/search/?q=CNP0000469 was used for alignment and data analysis. Short reads from each of the 124 BC1 samples, combined two replicates of parental lines ECD11 and T010000DH3, were aligned to the reference genome sequence. GBS-SNP-CROP pipeline v3.0 (Melo et al. 2016) was used to get SNP panel using Linux platform of AAFC Biocluster, following steps were performed: (1) parse the raw reads, (2) trim based on quality and adaptors, (3) demultiplex, (4) align with BWA-MEM and process with samtools, (5) parse mpileup outputs and produce the variant discovery matrix, (6) filter variants and call genotypes, and (7) create input files for downstream analyses. The trimming of the reads was performed using trimmomatic-0.39 (Bolger et al. 2014) prior to the read mapping. Trimming parameters were used as follows: data type = paired-end, number of threads = 10, phred quality score = 33, Trimmomatic ILLUMINACLIP string = TruSeq3-PE.fa:2:30:10:8:true, Trimmomatic LEADING value = 30, Trimmomatic SLIDINGWINDOW value 4:30, Trimmomatic TRAILING value = 30, Trimmomatic MINLEN value = 32. Identification of DNA variants (SNPs and InDels) in the DNA sequences each BC1 sample relative to the reference genome sequence was performed using the GBS-SNP-CROP pipeline v3.0, but only SNPs were used for further study. SNPs were filtered with the following parameters: mnHoDepth0 = 5, mnHoDepth1 = 20, mnHetDepth = 3, altStrength = 0.96, mnAlleleRatio = 0.25, mnCall = 0.75, mnAvgDepth = 3 and mxAvgDepth = 200. SNP panel from the 124 BC1 plants along with the parental lines ECD11 and T010000DH3 were used to get polymorphic sites in TASSEL5 (Bradbury et al. 2007). KNNimp imputation was done to recover missing data. Data were filtered with minimum count 5, minimum frequency 0.1 (10%), maximum frequency 1.0 (100%). SNP alleles from the susceptible parent (T010000DH3) were scored as ‘a’, resistant parent (ECD11) were scored as ‘b’ and heterozygous allele as ‘h’. As the parent T010000DH3 is a DH line, all SNP sites should theoretically be homozygous (a). Excel sort function was used to filter out allele ‘h’ from the DH line and SNP with higher missing sites.
Construction of linkage map and QTL mapping
To remove redundant markers, another two-step filtering was performed with JoinMap and the BIN function with ICIMapping. The high-quality polymorphic SNPs were further analyzed using JoinMap 4.1 (Van Ooijen 2011). Maximum likelihood in Kosambi’s model with a minimum logarithm of the odds (LOD) values of 10 was used to determine marker orders and positions in the genetic map. Only SNP sites that could be assigned into the 9 chromosomes of the C-genome sequence at LOD scores of 10.0 were kept. The set of filtered SNP sites obtained from JoinMap4.1 was used for binning of redundant markers, construction of linkage map, and mapping of QTLs for resistance to clubroot using the QTL IciMapping Inclusive Composite Interval Mapping (ICIM) method (Meng et al. 2015). Mapchart 2.1 (Voorrips 2002) was used to draw a linkage map based on the genetic location determined with QTL IciMapping. The LOD score threshold was set using a 1,000-permutation test with a Type I error of 0.05 for QTL declaration. The QTL effects were estimated as phenotypic variation explained (PVE) and additive (Add) values by each QTL.
Identification of potential candidate genes in the flanking region of QTL mapping
Coding sequences (CDS) of the genes in the identified QTL flanking regions were extracted from the reference genome sequence of B. oleracea ‘D134’ to perform gene annotation with Blast2GO (Conesa et al. 2005). Genes related to disease resistance and defense responses were identified from Blast2GO description. CDS of the disease resistance genes identified from Blast2GO were used for blast search at www.arabidopsis.org to find Arabidopsis homolog genes and the class of disease resistance proteins.
Search for A-genome syntenic regions of identified C-genome QTLs
The B. rapa reference genome sequence v3.0 (Zhang et al. 2018) downloaded from http://brassicadb.org/brad/downloadOverview.php were used to find C-genome synteny of identified QTL. Homolog of B. oleracea disease resistance genes to B. rapa and Arabidopsis genes were identified by blast search in http://brassicadb.cn/#/BLAST/ and http://www.arabidopsis.org/, respectively.
Results
Resistance to clubroot in the B. oleracea parental lines, F1 and BC1S1 population
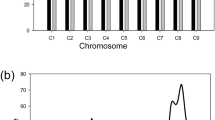
The clubroot reaction in the parental lines (ECD011 and T010000DH3), F1, controls and BC1 population was assessed against 10 pathotypes; 3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O as classified on the CCD set (Strelkov et al. 2018) (Fig. 1, Supplementary Table S1). ECD11 was highly or moderately resistant to all pathotypes (0–47.2% DSI), T010000DH3 was susceptible (66.7–94.4% DSI) for the ten pathotypes. The susceptible control ECD05 was highly susceptible to all the pathotypes (100% DSI). The F1 plants from the interspecific crosses T010000DH3 × ECD11 were highly susceptible (66.7–100% DSI) to 8 pathotypes; 3A, 2B, 5C, 3D, 5G, 3H, 5K and 3O, moderately resistant to 8J and 5L (33.3–50.0% DSI) (Supplementary Table S1). The frequency distribution in the BC1S1 population for resistance to all pathotypes showed continuous segregation patterns (Fig. 1). More BC1S1 lines with DSI distributed at 50–70% for pathotypes 3A, 2B, 3D, 70–90% for pathotypes 5C, 5G, 3H and 3O, 30–50% for 5L, 90–100% for 5K and 100% for 8J. Correlation coefficients among the disease severity index values for the 10 pathotypes ranged from 0.24 to 0.77, but all were significant at P < 0.01 except 5K at P < 0.05 (Table 1).
Alignment of GBS short reads into the Brassica oleracea reference genome sequence of cabbage DH line ‘D134’
In total 980.0 million (M) short reads were recovered from Illumina HiSeq2500 pair end sequencing from two replicates of parental lines and 124 BC1 lines (Supplementary Table S2). About 17.0 M and 15.4 M short reads were obtained from the parental lines ECD11 and T010000DH3, from there 16.1 M (94.7%) and 14.2 M (92.4%) reads were aligned into the B. oleracea reference genome sequence. A total 947.6 M short read sequence from 124 BC1 lines were identified ranging 0.9–13.9 M/line, average 7.6 M/line. The average number of reads aligned into the reference genome sequence from each line was 6.9 M, ranging 0.8–12.6 M and 84.4–93.9% (Supplementary Table S2).
Variants calling and polymorphic SNP selection
Total 8092 SNPs from 124 BC1 plants were assigned to nine chromosomes of the reference genome sequence. The average number of SNP/chromosome was 899, ranging 646–1129 of the reference genome sequence ‘D134’(Supplementary Table S3). The number of SNPs identified in the population was strongly correlated (R2 = 0.84) with length of chromosomes in the reference genome sequence of ‘D134’ (Supplementary Table S3). Total 7405 and 8011 SNPs from the parental lines ECD11 and T010000DH3 were assigned to the 9 chromosomes (Supplementary Table S3). The number of SNPs identified in the parental lines ECD11 and T010000DH3 was strongly correlated (R2 = 0.82, 0.84) with length of chromosomes in the reference genome sequence (Supplementary Table S3). Total 4,769 polymorphic SNP sites aligned to 9 chromosomes of ‘D134’ (Supplementary Table S3) and the number of polymorphic SNPs identified in the population were also strongly correlated (R2 = 0.71) with length of chromosomes (Supplementary Table S3). Total number SNPs and polymorphic SNPs were also positively correlated (R2 = 0.65) (Supplementary Table S3).
Construction of linkage map with comparison previously published map
A total of 1087 SNPs were filtered out in JoinMap and 2268 SNPs were filtered out by BIN function in ICIMapping. A genetic map consisting of 1,256.5 cM (Fig. S2) were constructed from the 1,414 non-redundant polymorphic SNP sites (Supplementary Table S3). The average number of SNPs mapped per chromosome was 157.1, ranging 126–185. The length of each chromosome ranged from 117.6 to 193.4 cM with an average length of 139 cM. The SNP interval of each chromosome ranged from 0.7 to 1.2 cM, with an average of 0.9 cM (Supplementary Table S3 and Table S4).
QTL mapping
QTL mapping was performed using the linkage map (Fig. S2) constructed with the 1414 SNP sites and %DSI values for resistance to 10 pathotypes (3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O). Seven QTLs were detected on chromosomes C01, C03, C04 and C08 (Fig. 2, Table 2). A single QTL for resistance to pathotype 2B was detected on chromosome C01, designated as Rcr_C01-1 with the peak between the SNP markers D134_C01_8,398,944 and D134_C01_7,492,399. QTLs for resistance to all the pathotypes were detected on chromosome C03. Rcr_C03-1 for resistance to 8 pathotypes; 2B, 5C, 5G, 3H, 8J, 5K, 5L and 3O (but not 3A and 3D) was located at the peak between the SNP markers D134_C03_9,211,088 and D134_C03_17,067,147. Rcr_C03-2 for resistance to pathotype 3D was identified at the peak between the SNP markers D134_C03_585,685 and D134_C03_1,026,778. Rcr_C03-3 with the peak between the SNP markers D134_C03_35,229,606 and D134_C03 _36,286,546, showed resistance to pathotype 3A. A single QTL was detected on chromosome C04, Rcr_C04-1 with the peak between the SNP markers D134_C04 _51,280,226 and D134_C04_52,146,319, showed resistance to pathotype 3D. Two QTLs were detected on chromosome C08; QTL Rcr_C08-1 with the peak between the SNP markers D134_C08_23,354,593 and D134_C08_24,782,998, showed resistance to pathotype 8J and 5K. Another QTL Rcr_C08-2 with the peak between the SNP markers D134_C08_28,507,471 and D134_C08_29,075,065, showed resistance to pathotype 5K.
LOD, PVE(%), Add values and confidence interval (CI) from the estimated QTL position varied among the QTLs, ranging from 3.2 to 23.8 for LOD, 8.3–65.6% for PVE, 6.1–39.4 for Add and 1–13 cM for CI (Fig. 2, Table 2). Among these seven QTLs, two QTLs in Rcr_C03-1 and Rcr_C08-1 were identified to resistance to eight and two pathotypes, respectively, with higher LOD value (4.2–23.8 and 4.7–16.4), PVE (16.0–65.6 and 8.3–23.5%) and Add values (14.3–41.4 and 14.7–30.3) (Table 2). The values of Add for five QTLs (Rcr_C01-1, Rcr_C03-1, Rcr_C03-2, Rcr_C03-3 and Rcr_C08-1) on C01, C03 and C08 were positive, indicating that the resistant loci were derived from the parent ECD11. However, QTLs (Rcr_C04-1, Rcr_C08-2) on C04, C08 were negative, indicating that the resistant loci were derived from the parent T010000DH3.
Identification of potential candidates genes for the QTLs
Genes in the flanking region of the seven QTLs were identified using CDS of the reference genome sequence of ‘D134’ by Blast2GO (Supplementary Table S5-8) and searched for candidate genes that encoded disease resistance proteins and defense-related genes (Table 3, Supplementary Table S9).
Rcr_C01-1 derived from T010000DH3 was identified in the flanking region of 7.49–8.39 Mb of chromosome C01 for resistance to pathotype 2B. One hundred eleven genes were identified in the flanking region of Rcr_C01-1, where 7 genes (Boc01g01056.1, Boc01g01060.1, Boc01g01022.1, Boc01g01023.1, Boc01g01024.1, Boc01g01063.1, Boc01g01078.1) encoded proteins with function related to plant defense response (Supplementary Table S5). Boc01g01060.1 is homologous to the Arabidopsis gene AT4G19510.2, which encoded TNL class protein, identified as possible candidate gene for Rcr_C01-1 (Table 3, Supplementary Table S9).
Rcr_C03-1 derived from ECD11 was identified in chromosome C03 in the flanking region of 9.21–17.06 Mb for resistance to 8 pathotypes (2B, 3H, 3O, 5C, 5G, 5K, 5L and 8J). Six hundred ninety genes were identified in the flanking region of Rcr_C03-1, where 10 genes (Boc03g00784.1, Boc03g00822.1, Boc03g00825.1, Boc03g00965.1, Boc03g00966.1, Boc03g00967.1, Boc03g00968.1, Boc03g00969.1, Boc03g00970.1 and Boc03g01016.1) encoded TNL disease resistance proteins (Supplementary Table S6). Boc03g00784.1 was homologous to the Arabidopsis gene AT2G14080.1. Two genes Boc03g00822.1 and Boc03g00825.1 were homologous to the Arabidopsis gene AT4G19510.1. Five genes Boc03g00965.1, Boc03g00966.1, Boc03g00967.1, Boc03g00968.1 and Boc03g00969.1 were homologous to the Arabidopsis gene AT4G36140.1. Boc03g00970.1 is homologous to the Arabidopsis gene AT4G36150.1. Boc03g01016.1 is homologous to the Arabidopsis gene AT3G44400.1 (Supplementary Table S9). These ten B. oleracea genes homologous to Arabidopsis genes encoding TNL class proteins could be the candidates for Rcr_C03-1.
Rcr_C03-2 derived from ECD11 was identified in chromosome C03 in the flanking region of 5.85–10.26 Mb for resistance to pathotype 3D. Forty-two genes were identified in the flanking region of Rcr_C03-2, where no gene encoded disease resistance proteins or function related to plant defense response (Supplementary Table S6).
Rcr_C03-3 derived from ECD11 was identified in chromosome C03 in the flanking region of 35.22–36.28 Mb, for resistance to pathotype 3A. Eighty-eight genes were identified in the flanking region of Rcr_C03-2, where no gene encoded disease resistance proteins or function related to plant defense response (Supplementary Table S6).
Rcr_C04-1 derived from T010000DH3 was identified in chromosome C04 in the flanking region of 51.28–52.14 Mb for resistance to pathotype 3D. Seventy-three genes were identified in the flanking region of Rcr_C04-1, where no gene encoded disease resistance protein or function related to plant defense response (Supplementary Table S7).
Rcr_C08-1 derived from ECD11 was identified in chromosome C08 in the flanking region of 23.35–24.78 Mb for resistance to pathotype 5K and 8J. Ninety-nine genes were identified in the flanking region of Rcr_C08-1, where 4 genes (Boc08g03058.1, Boc08g03059.1, Boc08g03179.1 and Boc08g03180.1) encoded disease resistance protein (Supplementary Table S8). Boc08g03058.1 and Boc08g03059.1 are homologous to Arabidopsis genes AT1G12290.1 and AT1G12220.2, both genes encoded CNL class proteins (Table 3, Supplementary Table S9). Another two genes Boc08g03179.1 and Boc08g03180.1 are homologous to Arabidopsis gene AT1G53350.1, which also encoded CNL class proteins (Table 3, Supplementary Table S9). These four B. oleracea genes Boc08g03058.1, Boc08g03059.1, Boc08g03179.1 and Boc08g03180.1 homologous to Arabidopsis genes AT1G12290.1, AT1G12220.2, AT1G53350.1 and AT1G53350.1, respectively, which encode CNL class proteins, could be possible candidate genes for Rcr_C08-1.
Rcr_C08-2 derived from T010000DH3 was identified in chromosome C08 in the flanking region of 28.50–29.07 Mb for resistance to pathotype 5K. Thirty-six genes were identified in the flanking region of Rcr_C08-2, where no gene encoded disease resistance protein or function related to plant defense response (Supplementary Table S8).
Polymorphic sequence variants identified in the QTLs flanking region between the two parents of chromosome C01, C03, C04 and C08 were searched. Out of 1139 genes in the flanking regions only 52 genes produced polymorphic variants (Supplementary Table S10).
Search for the syntenic regions of the two significant QTLs of C03 and C08 in the B. rapa ‘Chiifu’ reference genome sequence
B. rapa is a major source of CR genes or QTLs for clubroot disease resistance in Brassica species. In this study, the BC1 population was developed from B. oleracea with seven QTLs identified. More importantly, two QTLs Rcr_C03-1 and Rcr_C08-1 were identified for resistance to 8 and 2 pathotypes, respectively. B. oleracea chromosomes C03 and C08 have higher synteny with B. rapa chromosomes A03 and A08, respectively (Perumal et al. 2020). Many CR genes have been identified in chromosomes A03 and A08 (Yu et al. 2021; Rahaman et al. 2022). Two QTLs Rcr_C03-1 and Rcr_C08-1 with TNL or CNL genes identified in this study were compared with A03 and A08 of B. rapa using B. rapa reference genome sequence ‘Chiifu’ v3.0 (Zhang et al. 2018).
There were 10 TNL genes in the Rcr_C03-1 regions of C03, nine genes were homologous to A03 genes of B. rapa ‘Chiifu’ v3.0 (Supplementary Table S9, S11). Boc03g00784.1 (9,533,165–9,534,856), Boc03g00822.1 (10,163,079–10,168,624), Boc03g00825.1 (10,212,132–10,216,729), Boc03g00965.1 (11,746,815–11,747,411), Boc03g00967.1 (11,747,913–11,748,485), Boc03g00968.1 (11,748,756–11,749,391), Boc03g00969.1 (11,749,509–11,749,942), Boc03g00970.1 (11,751,167–11,755,194) and Boc03g01016.1 (12,152,885–12,168,584) were homologous to B. rapa A03 genes BraA03g049830.3C (25,366,429–25,366,902), BraA03g048820.3C (24,717,736–24,720,078), BraA03g048820.3C (24,717,736–24,720,078), BraA03g056630.3C (29,584,377–29,585,819), BraA03g048830.3C (24,729,618–24,732,164), BraA03g016950.3C (7,879,277–7,891,060) BraA03g051100.3C (26,191,764–26,194,343), BraA03g017700.3C (8,271,506–8,271,700), and BraA03g009600.3C (4,126,964- 4,129,030), respectively. There were 4 CNL genes identified in the Rcr_C08-1 regions of C08 (Table S9, S11). Boc08g03058.1 (23,486,420–23,486,905) and Boc08g03059.1 (23,487,114–23,487,434) were homologous to a single B. rapa A08 gene BraA08g031100.3C (21,138,657–21,138,929), while Boc08g03179.1 (24,623,210–24,624,463) and Boc08g03180.1 (24,625,065–24,626,401) were homologous to a single B. rapa A08 gene BraA08g032140.3C (21,640,250–21,641,322). Seventy-three SNP markers identified in the flanking region of C01, C03 and C08 of B. oleracea reference genome sequence of cabbage DH line D134, which can be used for marker-assisted selection (Table S12).
Discussion
Two major QTLs, Rcr_C03-1 and Rcr_C08-1, and five minor QTLs, Rcr_C01-1, Rcr_C03-2, Rcr_C03-3, Rcr_C04-1 and Rcr_C08-2, were detected from the current study on four chromosomes C01, C03, C04 and C08 against 10 pathotypes; 3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O. Previous QTLs against different pathotypes were identified from different B. oleracea resistant sources on those chromosomes of C01, C03, C04 and C08 (Voorrips et al. 1997; Moriguchi et al. 1999; Rocherieux et al. 2004; Nagaoka et al. 2010; Lee et al. 2016; Peng et al. 2018). Farid et al. 2020 also identified QTLs on C03, C04 and C08 with two pathotypes, 3A and 5X-LG2 through the association mapping. In this study, ten pathotypes were used for identification of QTLs in B. oleracea through the bi-parental mapping method. As there was no common reference genome sequence and molecular markers that can be used for the identification of QTLs, it was not possible to find relationships of the QTLs identified in this study with those previously identified by others.
Due to the rapid emergence of new pathotypes and the A-genome CR resistance breakdown in 1st generation CR cultivars, it becomes an urgency to utilize polygenic resistance of B. oleracea. Until now, most of the CR genes identified in Canada from A-genome of B. rapa (Yu et al. 2017, 2016, 2021; Chu et al. 2014; Huang et al. 2017; Gao et al. 2014; Karim et al. 2020; Rahaman et al. 2022) and B. napus (Fredua-Agyeman et al. 2016; Hasan et al. 2016; Zhang et al. 2016). One gene on chromosome A03 was used for developing the 1st generation CR cultivars in Canada. Recently, a report has been published on CR resistance of C-genome for resistance to pathotype 3 and 5X-LG2 (Dakouri et al. 2018). The study was based on RNA-seq. Compared to RNA-seq, QTL mapping from BC1/BC1S1 lines could be used for identifying CR loci for multiple pathotypes effectively (Yu et al. 2017). Therefore, identification of CR resistance from C-genome with more pathotypes became a priority to expand B. napus CR resistance from C-genome.
In this study, we performed bi-parental QTL mapping with polymorphic SNPs through GBS from 124 BC1 lines using a latest reference genome sequence ‘D134’. Depending on different morphotypes in B. oleracea species, five reference assemblies were published (Parkin et al. 2014; Liu et al. 2014; Sun et al. 2019; Belser et al. 2018; Lv et al. 2020) as variability between different morphotypes is high. Two types of DNA sequencing technologies were used: short-read technology such as next-generation sequencing (NGS) and long-read technology such as Oxford Nanopore Technology (ONT) and Pacific Biosciences (PacBio). Two B. oleracea genome sequences available based on NGS technology: the TO1000 (kale-like; B. oleracea var. alboglabra) assembly (Parkin et al. 2014) and the 02–12 (cabbage; B. oleracea var. capitata) assembly (Liu et al. 2014), but their errors and gaps make them difficult to use for many studies (Lee et al. 2016; Liu et al. 2016, 2017; Zhang et al. 2015). Another three B. oleracea genomes recently published based on combination of short and long-read technologies; the C-8 (cauliflower; B. oleracea L. var. botrytis) assembly (Sun et al. 2019), the HDEM (Broccoli; B. oleracea L. var. italica) assembly (Belser et al. 2018) and the ‘D134’ (Cabbage; B. oleracea var. capitata) assembly (Lv et al. 2020). Long-read technology with using efficient algorithms can provide high-quality assemblies in chromosome-level (Jiao et al. 2017; Belser et al. 2018; Michael et al. 2018). As the CR parental line ECD11 is a cabbage cultivar, we used ‘D134’ reference genome sequence (Lv et al. 2020) as the template for SNP calling in this study.
The number of SNPs site per chromosome is often correlated with chromosome size (Yu et al. 2016). In this study, the number of SNPs and polymorphic SNPs identified in the population were strongly correlated (R2 = 0.84, 0.71) of the reference genome sequence (Table S3). The total numbers of SNPs shows a better correlation with the chromosome length than the number of polymorphic SNPs, suggesting that polymorphic SNPs may be clustered. However, this may not be an issue for filtering, construction of genetic map and subsequent QTL mapping as BIN function was performed to remove redundant SNP loci using IciMapping. Although polymorphic SNPs seem more clustered but still total number SNPs and polymorphic SNPs were also positively correlated (R2 = 0.65). Genetic linkage map is pre-requisite for QTL analysis and genetic mapping. Linkage map length could depend on population type and size, reference genome sequence size as well as number of polymorphic variants used for mapping. In this study, we constructed the linkage map with polymorphic variants of 124 BC1 lines using the latest reference genome sequence ‘D134’. It was 1,256.5 cM in length, longer than previously published B. oleracea linkage maps; 879.9 cM (Lee et al. 2016), 913.5 cM (Haque et al. 2017) and 1028.0 cM (Peng et al. 2018).
The R parental line ECD11 was moderate to highly resistant to all 10 pathotypes, while S parental line T010000DH3 was highly susceptible for all pathotypes. The F1 plants from the interspecific crosses T010000DH3 × ECD11 were moderate to highly susceptible to all pathotypes and BC1S1 population showed continuous distribution of %DSI (Fig. 1), which indicate that CR resistance of these 10 pathotypes is controlled by QTLs. Similar quantitative resistance was reported in B. oleracea for clubroot (Figdore et al. 1993; Nagaoka et al. 2010) as well black rot (Tonu et al. 2013) diseases. The frequency distribution in the BC1S1 population for resistance to some pathotypes also showed skewed tendency, similar skewed phenotype also reported for clubroot quantitative resistance in B. oleracea (Figdore et al. 1993; Peng et al. 2018; Ce et al. 2021). These results indicated that ECD11 clubroot resistance is controlled by quantitative manner with multiple genes effect.
A QTL that can be consistently detected with a PVE of > 10% of trait value can be designated as the main effect QTL or major QTL (Wang et al. 2019). In this study, one QTL, Rcr_C03-1 on C03 which contributed 16.0–65.6% phenotypic variation for 8 pathotypes; 2B, 5C, 5G, 3H, 8J, 5K, 5L and 3O (Table 2). Another QTL, Rcr_C08-1 were identified with 8.3 and 23.5% PVE for resistance to 8J and 5K, respectively (Table 2). As these two QTLs were identified for resistance to more than one pathotypes with PVEs of > 10% in the most cases. They can consider as major QTLs in this study. These two QTLs in Rcr_C03-1 and Rcr_C08-1 were identified with higher LOD value, PVE and Add values, indicating that they might have significant contribution for CR resistance in ECD11. Previously, one major QTL (Pb-Bo1) was detected on C01 for five isolates (Pathotypes 1, 2, 4 & 7; according to the classification by Some et al. 1996) and explained 20.7–80.7% of the phenotypic variation (Rocherieux et al. 2004). Five minor QTLs Rcr_C01-1, Rcr_C03-2, Rcr_C03-3, Rcr_C04-1 and Rcr_C08-2 were detected on chromosomes C01, C03, C04 and C08 which contributed 8.3–18.7% PVE for only one pathotype each (Table 2). A previous study reported that a single involvement of the major CR gene, or accumulation of CR genes in the minor CR–QTL, is not enough to confer sufficient resistance (Tomita et al. 2013). Further investigations for understanding individual or cumulative effect of identified major QTLs in ECD11 are needed. The values of Add for the five QTLs (Rcr_C01-1, Rcr_C03-1, Rcr_C03-2, Rcr_C03-3, Rcr_C08-1) on C01, C03 and C08 were positive, while for 2 QTLs (Rcr_C04-1 and Rcr_C08-2) on C04 and C08 were negative, indicating that the five resistant loci were derived from the resistant parent ECD11 and two resistant loci were derived from the susceptible parent T010000DH3. The presence of QTL for resistance to clubroot in T010000DH3 needs further confirmation. Previously, a resistant locus, PbBo(GC)1 identified from susceptible broccoli parent account for 9% of the phenotypic variation (Nagaoka et al. 2010).
Among identified 7 CR QTLs, disease resistance proteins and defense-related genes were identified in three QTLs (Rcr_C01-1, Rcr_C03-1 and Rcr_C08-1) (Table 3). From the Rcr_C01-1 flanking region, a potential candidate TNL gene Boc01g01060.1 was identified. From the Rcr_C03-1 flanking region, ten TNL genes (Boc03g00784.1, Boc03g00822.1, Boc03g00825.1, Boc03g00965.1, Boc03g00966.1, Boc03g00967.1, Boc03g00968.1, Boc03g00969.1, Boc03g00970.1 and Boc03g01016.1) encoding disease resistance proteins were identified, which were homologous to five Arabidopsis genes. Two genes Boc03g00822.1 and Boc03g00825.1 were homologous to AT4G19510.1 and five genes Boc03g00965.1, Boc03g00966.1, Boc03g00967.1, Boc03g00968.1 and Boc03g00969.1 were homologous to AT4G36140.1. Identified candidate CR genes of B. oleracea which are homologous to the same Arabidopsis gene producing similar resistance proteins or not are to be determined. From the Rcr_C08-1 flanking region, 4 genes (Boc08g03058.1, Boc08g03059.1, Boc08g03179.1 and Boc08g03180.1) encoding disease resistance proteins were identified, which were homologous to 3 Arabidopsis genes and produce CNL class proteins. TNL genes are rare in monocotyledons, but CNL genes are quite common in both monocotyledons and dicotyledons (Tarr et al. 2009; Liu et al. 2021). Although previously three genes CRa, Crr1a and CRb have been cloned and TNL class proteins were reported for clubroot resistance from dicot Brassica. (Ueno et al. 2012; Hatakeyama et al. 2013, 2017), there is no report on CNL class proteins for resistance to clubroot.
In this study, CR QTLs with disease resistant genes were identified in chromosomes C01, C03 and C08 from ECD11. B. oleracea chromosomes C03 and C08 have higher synteny with B. rapa chromosome A03 and A08 (Perumal et al. 2020). In addition, A03 and A08 contain many CR genes. Therefore, the identified major QTLs of C03 and C08 with candidate disease resistant genes in this study were compared with A03 and A08 of the B. rapa reference genome sequence ‘Chiifu’ v3.0 (Zhang et al. 2018) (Supplementary Table S11). For Rcr_C03-1, among ten candidate genes of C03, four genes, Boc03g00784.1, Boc03g00822.1, Boc03g00825.1 and Boc03g00967.1, were homologous to B. rapa A03 genes BraA03g049830.3C (25,366,429–25,366,902), BraA03g048820.3C (24,717,736–24,720,078), BraA03g048820.3C (24,717,736–24,720,078) and BraA03g048830.3C (24,729,618–24,732,164), which resides in the flanking region of previously identified CR major gene Rcr1 (24.57–25.71 Mb) and Rcr4 region (23.82–26.08 Mb). One gene, Boc03g00969.1, was homologous to B. rapa A03 gene BraA03g051100.3C (26,191,764–26,194,343), resides in the flanking region of previously identified QTL, Rcr4 region (23.82–26.08 Mb). Rest of the four genes, Boc03g00965.1, Boc03g00968.1, Boc03g00970.1 and Boc03g01016.1, were homologous to B. rapa A03 genes BraA03g056630.3C (29,584,377–29,585,819), BraA03g016950.3C (7,879,277–7,891,060), BraA03g017700.3C (8,271,506–8,271,700) and BraA03g009600.3C (4,126,964–4,129,030) accordingly. According to our knowledge, no CR genes were reported previously for these four genes, so these four genes could be considered as novel candidate genes in the cluster for CR resistance. For Rcr_C08-1, four candidate resistant genes were identified. Boc08g03058.1 and Boc08g03059.1 were homologous to a single B. rapa A08 gene BraA08g031100.3C (21,138,657–21,138,929), while Boc08g03179.1 and Boc08g03180.1 were homologous to a single B. rapa A08 gene BraA08g032140.3C (21,640,250–21,641,322). Polymorphic sequence variants in the QTLs flanking regions were also searched between the two parents (Supplementary Table S10). Out of 1,139 genes in the flanking region only 52 genes produced polymorphic variants. Even though higher coverage of GBS in the parents, only two defense-related genes (Boc01g01078.1 and Boc03g00965.1) produced polymorphic variants. Although the genome sequence of susceptible line DH3 is available, whole genome sequencing of resistant line ECD11 is needed so comparison of two genome sequences in the target regions could be performed. Cloning of the genes is also required for confirming the CNL gene involvement for CR resistance. This study focuses on identification of QTLs and mapping QTLs into the respective genomic regions. There are genes encoding disease resistance proteins in some of the QTL intervals. However, further study will be needed to determine if they confer tolerance or resistance. The comparison of previously identified QTLs on C01, C03, C08 was not possible due to lack of common molecular markers, reference genome and different source of clubroot pathogen pathotypes used in those studies. A total 15 genes encoding TNL/CNL class disease resistance proteins were identified in the Rcr_C01-1, Rcr_C03-1 and Rcr_C08-1 flanking regions for resistance to 10 pathotypes. Identified CR QTLs from B. oleracea cultivar ECD11 can be transferred to B. napus either interspecific breeding or developing new re-synthesized B. napus, which could significantly contribute to development of new CR canola varieties against new races of P. brassicae in Western Canada.
Data availability
The raw datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information (NCBI) repository, BioProject accession: PRJNA948018. Data will be released on 31 July, 2023, or upon manuscript publication. https://www.ncbi.nlm.nih.gov/sra/PRJNA948018.
References
Belser C, Istace B, Denis E, Dubarry M, Baurens FC, Falentin C et al (2018) Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plant 4:879–887. https://doi.org/10.1038/s41477-018-0289-4
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Cao T, Manolii VP, Zhou Q, Hwang SF, Strelkov SE (2020) Effect of canola (Brassica napus) cultivar rotation on Plasmodiophora brassicae pathotype composition. Can J Plant Sci 100(2):218–225. https://doi.org/10.1139/cjps-2019-0126
Ce F, Mei J, He H, Zhao Y, Hu W, Yu F, Li Q, Ren X, Si J, Song H, Qian W (2021) Identification of candidate genes for clubroot-resistance in Brassica oleracea using quantitative trait loci-sequencing. Front Plant Sci 12:703520. https://doi.org/10.3389/fpls.2021.703520
Chu M, Song T, Falk KC, Zhang X, Liu X, Chang A (2014) Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15(1):1166. https://doi.org/10.1186/1471-2164-15-1166
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Dakouri A, Zhang X, Peng G, Falk KC, Gossen BD, Strelkov SE, Yu F (2018) Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci Rep 8:17657. https://doi.org/10.1038/s41598-018-36187-5
Dakouri A, Lamara M, Karim MM, Wang J, Chen Q, Gossen BD et al (2021) Identification of resistance loci against new pathotypes of Plasmodiophora brassicae in Brassica napus based on genome-wide association mapping. Sci Rep 11:6599. https://doi.org/10.1038/s41598-021-85836-9
Dias JS, Ferreira ME, Williams PH (1993) Screening of Portuguese cole landraces (Brassica oleacea L.) with Peronospora parasitica and Plasmodiophora brassicae. Euphytica 67:135–141. https://doi.org/10.1007/BF00022736
Diederichsen E, Frauen M, Linders EGA, Hatakeyama K, Hirai M (2009) Status and perspectives of clubroot resistance breeding in crucifer crops. J Plant Growth Regul 28:265–281. https://doi.org/10.1007/s00344-009-9100-0
Dixon GR (2009) The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J Plant Growth Regul 28:194–202. https://doi.org/10.1007/s00344-009-9090-y
Donald EC, Porter IJ (2014) Clubroot in Australia: The history and impact of Plasmodiophora brassicae in Brassica crops and research efforts directed towards its control. Can J Plant Pathol 36:66–84. https://doi.org/10.1080/07060661.2013.873482
Engqvist LG (1994) Distribution of clubroot (Plasmodiophora brassicae Wor) in Sweden and the effect of infection on oil content of oilseed rape (Brassica napus L.). J Swedish Seed Assoc 104(2):82–86
Farid M, Yang RC, Kebede B, Rahman H (2020) Evaluation of Brassica oleracea accessions for resistance to Plasmodiophora brassicae and identification of genomic regions associated with resistance. Genome 63(2):91–101. https://doi.org/10.1139/gen-2019-0098
Figdore SS, Ferrerira ME, Slocum MK, Williams PH (1993) Association of RFLP markers with trait loci affecting clubroot resistance and morphological characters in Brassica oleracea L. Euphytica 69:33–44. https://doi.org/10.1007/BF00021723
Fredua-Agyeman R, Rahman MH (2016) Mapping of the clubroot disease resistance in spring Brassica napus canola introgressed from European winter canola cv. ‘Mendel.’ Euphytica 211:201–213. https://doi.org/10.1007/s10681-016-1730-2
Fredua-Agyeman R, Hwang SF, Strelkov SE, Zhou Q, Feindel D (2018) Potential loss of clubroot resistance genes from donor parent Brassica rapa subsp. Rapifera (ECD 04) during doubled haploid production. Plant Pathol 67(4):892–901. https://doi.org/10.1111/ppa.12816
Gao F, Hirani AH, Liu J, Liu Z, Fu G, Wu C et al (2014) Fine mapping a clubroot resistance locus in Chinese cabbage. J Amer Soc Hort Sci 139:247–252
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923
Gossen BD, Adhikari KK, McDonald MR (2012) Effect of seeding date on development of clubroot in short-season Brassica crops. Can J Plant Pathol 34:516–523. https://doi.org/10.1080/07060661.2012.722129
Hasan MJ, Rahman H (2016) Genetics and molecular mapping of resistance to Plasmodiophora brassicae pathotypes 2, 3, 5, 6, and 8 in rutabaga (Brassica napus var. napobrassica). Genome 59(10):805–815. https://doi.org/10.1139/gen-2016-0034
Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, Matsumoto S (2013) Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE 8:e54745. https://doi.org/10.1371/journal.pone.0054745
Hatakeyama K, Niwa T, Kato T, Ohara T, Kakizaki T, Matsumoto S (2017) The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol Genet and Genomics 292(2):397–405. https://doi.org/10.1007/s00438-016-1281-1
Hollman KB, Hwang SF, Manolii VP, Strelkov SE (2021) Pathotypes of Plasmodiophora brassicae collected from clubroot resistant canola (Brassica napus L.) cultivars in western Canada in 2017–2018. Can J Plant Pathol 43(4):622–630. https://doi.org/10.1080/07060661.2020.1851893
Hoque M, Shea DJ, Asada M, Doullah MAU, Shimizu M, Fujimoto R et al (2017) QTL mapping for tuberous stem formation of kohlrabi (Brassica oleracea var. gongylodes L.). Mol Breeding 37:109. https://doi.org/10.1007/s11032-017-0709-6
Horiuchi S, Hori M (1980) A simple greenhouse technique for obtaining high levels of clubroot incidence. Bull Chugoku Natl Agric Exp 17:33–55
Huang Z, Peng G, Liu X, Deora A, Falk KC, Gossen BD et al (2017) Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front Plant Sci 8:1448. https://doi.org/10.3389/fpls.2017.01448
Hwang SF, Howard RJ, Strelkov SE, Gossen BD, Peng G (2014) Management of clubroot (Plasmodiophora brassicae) on canola (Brassica napus) in western Canada. Can J Plant Pathol 36:49–65. https://doi.org/10.1080/07060661.2013.863806
Jiao WB, Accinelli GG, Hartwig B, Kiefer C, Baker D, Severing E et al (2017) Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res 27:778–786
Karim MM, Dakouri A, Zhang Y, Chen Q, Peng G, Strelkov SE (2020) Two clubroot-resistance genes, Rcr3 and Rcr9wa, mapped in Brassica rapa using bulk segregant RNA sequencing. Int J Mol Sic 21(14):5033. https://doi.org/10.3390/ijms21145033
Kuginuki Y, Hiroaki Y, Hirai M (1999) Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot-resistant cultivars of Chinese cabbage (Brassica rapa L. spp. pekinensis). Eur J Plant Pathol 105:327–332. https://doi.org/10.1023/A:1008705413127
Lee J, Izzah NK, Choi BS, Joh HJ, Lee SC, Perumal S et al (2016) Genotyping-by-sequencing map permits identification of clubroot resistance QTLs and revision of the reference genome assembly in cabbage (Brassica oleracea L.). DNA Res 23(1):29–41. https://doi.org/10.1093/dnares/dsv034
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930. https://doi.org/10.1038/ncomms4930
Liu Y, Li D, Yang N, Zhu X, Han K, Gu R, Bai J, Wang A, Zhang Y (2021) Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in Chinese cabbage. Int J Mol Sci 22(8):4266. https://doi.org/10.3390/ijms22084266
Lv H, Wang Y, Han F, Ji J, Fang Z, Zhuang M et al (2020) A high-quality reference genome for cabbage obtained with SMRT reveals novel genomic features and evolutionary characteristics. Sci Rep 10:12394. https://doi.org/10.1038/s41598-020-69389-x
Manzanares-Dauleux MJ, Divaret I, Baron F, Thomas G (2000) Evaluation of French Brassica oleracea landraces for resistance to Plasmodiophora brassicae. Euphytica 113:211–218. https://doi.org/10.1023/A:1003997421340
McHale L, Tan X, Koehl P, Michelmore R (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212. https://doi.org/10.1186/gb-2006-7-4-212
Melo AT, Bartaula R, Hale I (2016) GBS-SNP-CROP: a reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinform 12(17):29. https://doi.org/10.1186/s12859-016-0879-y
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283. https://doi.org/10.1016/j.cj.2015.01.001
Michael TP, Jupe F, Bemm F, Motley ST, Sandoval JP, Lanz C et al (2018) High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell. Nat Commun 9:541. https://doi.org/10.1038/s41467-018-03016-2
Moriguchi K, Kimizuka-Takagi C, Ishii K, Nomura K (1999) A genetic map based on RAPD, RFLP, Isozyme, morphological markers and QTL analysis for clubroot resistance in Brassica oleracea. Breed Sci 49:257–265. https://doi.org/10.1270/jsbbs.49.257
Morinaga T (1934) Interspecific hybridization in Brassica VI. The cytology of F1 hybrids of B. juncea and B. nigra. Cytologia 6:62–67
Nagaharu U, Nagaharu N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Nagaoka T, Doullah MAU, Matsumoto S, Kawasaki S, Ishikawa T, Hori H, Okazaki K (2010) Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theor Appl Genet 120:1335–1346. https://doi.org/10.1007/s00122-010-1259-z
Pageau D, Lajeunesse J, Lafond J (2006) Impact del’hernie des crucifères [Plasmodiophora brassicae] sur la productivité et la qualité du canola. Can J Plant Pathol 28(1):137–143. https://doi.org/10.1080/07060660609507280
Parkin IA, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77. https://doi.org/10.1186/gb-2014-15-6-r77
Peng G, Lahlali R, Hwang SF, Pageau D, Hynes RK, McDonald MR (2014) Crop rotation, cultivar resistance, and fungicides/biofungicides for managing clubroot (Plasmodiophora brassicae) on canola. Can J Plant Pathol 36(Suppl. 1):99–112. https://doi.org/10.1080/07060661.2013.860398
Peng L, Zhou L, Li Q, Wei D, Ren X, Song H, Mei J, Si J, Qian W (2018) Identification of quantitative trait loci for clubroot resistance in Brassica oleracea with the use of Brassica SNP microarray. Front Plant Sci 9:822. https://doi.org/10.3389/fpls.2018.00822
Perumal S, Koh CS, Jin L, Buchwaldt M, Higgins EE, Zheng C, Sankoff D, Robinson SJ, Kagale S, Navabi ZK (2020) A high-contiguity Brassica nigra genome localizes active centromeres and defines the ancestral Brassica genome. Nat Plants 6:929–941. https://doi.org/10.1038/s41477-020-0735-y
Rahaman M, Strelkov SE, Hu H, Gossen B, Yu F (2022) Identification of a genomic region containing genes involved in resistance to four pathotypes of Plasmodiophora brassicae in Brassica rapa turnip ECD02. The Plant Genome 15:e20245. https://doi.org/10.1002/tpg2.20245
Rahman H, Peng G, Yu F, Falk KC, Kulkarni M, Selvaraj G (2014) Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Can J Plant Pathol 36(Suppl. 1):122–134. https://doi.org/10.1080/07060661.2013.862571
Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux MJ (2004) Isolate-specific and broad spectrum QTLs are involved in the control in Brassica oleracea. Theor Appl Genet 108(8):1555–1563. https://doi.org/10.1007/s00122-003-1580-x
Sedaghatkish A, Gossen BD, Yu F, Torkamaneh D, McDonald MR (2019) Whole-genome DNA similarity and population structure of Plasmodiophora brassicae strains from Canada. BMC Genomics 20(1):744. https://doi.org/10.1186/s12864-019-6118-y
Some A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F (1996) Variation for virulence on Brassica napus L. among Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol 45:432–439. https://doi.org/10.1046/j.1365-3059.1996.d01-155.x
Strelkov SE, Hwang SF, Manolii VP, Cao T, Feindel D (2016) Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur J Plant Pathol 145:517–529. https://doi.org/10.1007/s10658-016-0888-8
Strelkov SE, Hwang SF, Manolii VP, Cao T, Fredua-Agyeman R, Harding MW, Peng G, Gossen BD, McDonald MR, Feindel D (2018) Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can J Plant Pathol 40:284–298. https://doi.org/10.1080/07060661.2018.1459851
Sun D, Wang C, Zhang X, Zhang W, Jiang H, Yao X et al (2019) Draft genome sequence of cauliflower (Brassica oleracea L. var. botrytis) provides new insights into the C genome in Brassica species. Hortic Res 6:82. https://doi.org/10.1038/s41438-019-0164-0
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M (2003) Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet 107:997–1002. https://doi.org/10.1007/s00122-003-1309-x
Tarr DEK, Alexander HM (2009) TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res Notes 2:197. https://doi.org/10.1186/1756-0500-2-197
Tewari JP, Strelkov SE, Orchard D, Hartman M, Lange RM, Turkington TK (2005) Identification of clubroot of crucifers on canola (Brassica napus) in Alberta. Can J Plant Pathol 27:143–144. https://doi.org/10.1080/07060660509507206
Tomita H, Shimizu M, Doullah MAU, Fujimoto R, Okazaki K (2013) Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Mol Breed 32:889–900. https://doi.org/10.1007/s11032-013-9918-9
Tonu NN, Doullah MAU, Shimizu M, Karim MM, Kawanabe T, Fujimoto R, Okazaki K (2013) Comparison of positions of QTLs conferring resistance to Xanthomonas campestris pv. campestris in Brassica oleracea. Amer J Plant Sci 4:11–20. https://doi.org/10.4236/ajps.2013.48A002
Ueno H, Matsumoto E, Aruga D, Kitagawa S, Matsumura H, Hayashida N (2012) Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol Biol 80(6):621–629. https://doi.org/10.1007/s11103-012-9971-5
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Gen Res 93:343–349. https://doi.org/10.1017/S0016672311000279
Voorrips RE (1995) Plasmodiophora brassicae: aspects of pathogenesis and resistance in Brassica oleracea. Euphytica 83:139–146. https://doi.org/10.1007/BF01678041
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Voorrips RE, Jongerius MC, Kanne HJ (1997) Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of doubled haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor Appl Genet 94:75–82. https://doi.org/10.1007/s001220050384
Wang L, Yang X, Cui S, Mu G, Sun X, Liu L (2019) QTL mapping and QTL × environment interaction analysis of multi-seed pod in cultivated peanut (Arachis hypogaea L.). Crop J 7:249–260. https://doi.org/10.1016/j.cj.2018.11.007
Xp L, Yang C, Fq H, Zy F, Lm Y, Zhuang M et al (2016) Genetics and fine mapping of a yellow-green leaf gene (ygl-1) in cabbage (Brassica oleracea var. capitata L.). Mol Breed 36:82. https://doi.org/10.1007/s11032-016-0509-4
Xp L, Bz G, Fq H, Zy F, Lm Y, Zhuang M et al (2017) Genetics and fine mapping of a purple leaf gene, BoPr, in ornamental kale (Brassica oleracea L. var. acephala). BMC Genom 18:230. https://doi.org/10.1186/s12864-017-3613-x
Yu F, Zhang X, Huang Z, Chu M, Song T, Falk KC (2016) Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS ONE 11:e0153218. https://doi.org/10.1371/journal.pone.0153218
Yu F, Zhang X, Peng G, Falk KC, Strelkov SE, Gossen BD (2017) Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci Rep 7:4516. https://doi.org/10.1038/s41598-017-04903-2
Yu F, Zhang Y, Wang J, Chen Q, Karim MM, Gossen BD, Peng G (2021) Identification of two major QTLs in Brassica napus lines with introgressed clubroot resistance from turnip cultivar ECD01. Front Plant Sci 12:785989. https://doi.org/10.3389/fpls.2021.785989
Zhang B, Liu C, Wang Y, Yao X, Wang F, Wu J, King GJ, Liu K (2015) Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol 206(4):1513–1526. https://doi.org/10.1111/nph.13335
Zhang H, Feng J, Hwang SF, Strelkov SE, Falax I, Sun R (2016) Mapping of clubroot (Plasmodiophora brassicae) resistance in canola (Brassica napus). Plant Pathol 65:435–440. https://doi.org/10.1111/ppa.12422
Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, Liang J, Cai C, Liu Z, Liu B (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Horticult Res 5:50. https://doi.org/10.1038/s41438-018-0071-9
Acknowledgements
We thank Dr. S.E. Strelkov at the University of Alberta for providing the strains of P. brassicae, Dr. G. R. Dixon (The University of Warwick) for proving the resistant parent and Dr. I. Parkin for providing the susceptible parent, F. Fu, A. Dakouri, M. Kehler, Y. Zhang, Q. Chen and J. Wang for their technical assistance.
Funding
Open Access funding provided by Agriculture & Agri-Food Canada. This work was funded by a competitive grant from the Canola Agri-Science Cluster. The funding body played no role in the design of the study or in the collection, analysis, and interpretation of the data or writing of the manuscript.
Author information
Authors and Affiliations
Contributions
MK designed and conducted all the experiments, performed data analysis and prepared the manuscript. FY conceived of this study, designed the experiments, collected materials and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Jacqueline Batley.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2023_4483_MOESM3_ESM.png
The linkage map of Brassica oleracea consisting of 1,414 SNP sites extracted from using Brassica oleracea reference genome sequence of cabbage doubled haploid line ‘D134’ (PNG 98 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Karim, M.M., Yu, F. Identification of QTLs for resistance to 10 pathotypes of Plasmodiophora brassicae in Brassica oleracea cultivar ECD11 through genotyping-by-sequencing. Theor Appl Genet 136, 249 (2023). https://doi.org/10.1007/s00122-023-04483-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04483-y