Abstract
Apomixis is an asexual mode of reproduction through seeds where progeny are clones of the mother plants. Naturally apomictic modes of reproduction are found in hundreds of plant genera distributed across more than 30 plant families, but are absent in major crop plants. Apomixis has the potential to be a breakthrough technology by allowing the propagation through seed of any genotype, including F1 hybrids. Here, we have summarized the recent progress toward synthetic apomixis, where combining targeted modifications of both the meiosis and fertilization processes leads to the production of clonal seeds at high frequencies. Despite some remaining challenges, the technology has approached a level of maturity that allows its consideration for application in the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The goal of engineered apomixis
In the last century, significant improvements in yield and other desirable crop traits were seen following the widespread adoption of hybrid crops, the product of F1 crosses between two high performing, divergent lines (Hochholdinger and Baldauf 2018). Hybrid vigor, or heterosis, is the observed higher performance of an F1 hybrid over that of either parent line; this performance is reduced following the random segregation of genetic information in subsequent generations. Utilization of hybrid vigor thus depends today on the production of F1 hybrids by crosses. Apomixis is a naturally occurring mode of reproduction in plants that forms seeds identical in genetic makeup to the maternal parent and represents an efficient means of clonal reproduction that could fix parental genotypes such as F1 hybrids. The occurrence of apomixis is, however, absent in modern crops and restricted to widely spread but majorly non-cultivated plant species. In this review we aim to summarize work to synthetically produce apomixis in economically relevant crop species by linking natural apomixis with engineering strategies, with a further outlook for its suitability in modern agriculture.
Sexual reproduction in flowering plants
Flowering plants have alternating phases, in which the sporophytic diploid phase (2n) alternates with the gametophytic haploid phase (n). Meiosis, occurring in both male and female organs, constitutes the transition from the sporophytic to the gametophytic phase. It produces haploid spores that divide mitotically to generate the male (pollen grain) and female (embryo sac). The gametophytic structures are composed of only a few cells and rely on the sporophyte for nourishment and development. In females, only one of the four spores survives and undergoes three mitotic divisions to form an eight-nuclei embryo sac. In the mature embryo sac, three cells are toward one side (micropylar, entry point of the pollen tube), the egg cell and two synergid cells, while three antipodal cells lie at the opposite side (chalazal end), while in between lies a double haploid (diploid) central cell that is produced by the fusion of two haploid cells (Skinner and Sundaresan 2018). In males, each of the four haploid cells survives and divides twice mitotically to generate a pollen grain that contains two sperm cells and one vegetative cell. At fertilization, the sperm cells are delivered by the pollen grain to the female gametophyte. One sperm cell fuses with the egg cell, resulting in a diploid embryo (Fig. 1B). The second sperm cell fuses with the diploid central cell, resulting in a triploid endosperm, the nourishing tissue of the embryo in the seed (Hafidh and Honys 2021).
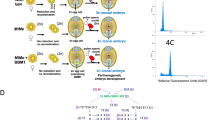
Engineering apomeiosis and parthenogenesis. Mutations in three meiotic genes (MiMe) alter crucial stages of meiosis to result in a mitotic-like division of chromosomes, mimicking and providing a tool to implement apomeiosis (A). Embryogenesis in many plants results from fertilization of the female-derived ovule and central cell by the male-derived pollen to give rise to a diploid zygote and triploid endosperm (B, left). Prior to fertilization, BBM1 and PAR are expressed in the male gamete; redirecting their expression to the ovule can result in the formation of haploid zygotes (B, right). Alternatively, mutations in MTL/PLA1/NLD, DMP, or CENH3 can hinder fertilization by disrupting one parental gamete contribution, and can produce haploid zygotes (C). By pairing MiMe with male expressed BBM1/PAR or mutations in MTL/PLA1/NLD, DMP, or CENH3, clonal progeny can be obtained that represent synthetic apomicts (D). Figure created with BioRender.com
Natural apomicts
Meiosis and fertilization are thus the two key steps of the sexual life cycle in flowering plants. Some plants, however, bypass meiosis and fertilization to reproduce asexually through seeds, a mode of reproduction known as apomixis. Apomixis (gametophytic apomixis) has been documented in about 400 species (Asker and Jerling 1992; Carman 1997; Kellogg 1990; Majeský et al. 2017) and distributed across more than 30 families (Table 1), but the majority of the species belong to three families: Poaceae, Asteraceae and Rosaceae. Adventitious embryony is mostly found in Orchidaceae and Rutaceae citrus plants (Asker and Jerling 1992; Carman 1997; Hand and Koltunow 2014; Hojsgaard et al. 2014).
Several types of natural apomixis have been defined, depending on the origin of the clonal embryo: gametophytic apomixis retains the development of an embryo sac and is further divided into diplospory and apospory, depending on the origin of this embryo sac (Koltunow and Grossniklaus 2003; Underwood and Mercier 2022). In diplospory, a modified female meiosis that resembles mitosis produces non-recombined, clonal diploid spores. This spore divides mitotically to form a mature embryo sac that cytologically appears similar to the wild type. The egg cell, which is thus diploid and clonal, enters embryogenesis without fertilization, a process known as parthenogenesis. In apospory, a somatic cell derived from the ovule develops directly into a diploid embryo sac. The presence of several embryo sacs or the absence of antipodal cells distinguishes apospory from other forms of apomixis (Conner and Ozias-Akins 2017; Koltunow 1994). Although each ovule frequently produces several aposporous embryo sacs, typically only one of these develops into an embryo by parthenogenesis. Another mode of apomixis, classified as sporophytic, is adventitious embryony, in which embryos develop directly from somatic cells of the ovule. Because sexual reproduction occurs in the same ovule in parallel to apomictic embryogenesis, polyembryonic seed development is very common in sporophytic apomixis.
On the basis of the penetrance of apomictic progeny formation, apomicts are classified into two groups: obligate apomicts and facultative apomicts. Obligate apomicts reproduce mostly through apomictic means, but a small fraction reproduces sexually, whereas facultative apomicts show the reverse (Asker and Jerling 1992; Mráz and Zdvořák, 2019).
A seed embryo is surrounded and fed by an endosperm, a tissue equivalent to the placenta in mammals. Endosperm development is as important as the embryo for the generation of viable seeds. In sexual reproduction, the endosperm is triploid as the result of the fusion of the female diploid central cell with one haploid sperm cell. In most apomictic plants, the embryo develops without fertilization (parthenogenesis), but for endosperm (see above), the central cell must be pollinated and fertilized, a process called pseudogamy (Hojsgaard and Hörandl 2019; Nogler 1984). Autonomous development of endosperm is also reported in a few apomicts that do not need fertilization (Koltunow 1994; Koltunow and Grossniklaus 2003).
Engineering apomixis step 1: apomeiosis
Despite being present in many species, apomixis is absent in major crops. In the last decade, much progress has been made in engineering de novo apomixis by tinkering with the process of sexual reproduction. The first crucial step is to induce apomeiosis, the modification of meiosis into a mitotic-like division. Three crucial events distinguish meiosis from mitosis: (i) recombination in prophase of meiosis I, (ii) co-segregation of sister chromatids at meiosis I, and (iii) a second round of division. Abolishing each of these differences with appropriate mutations can turn meiosis into mitosis as a strategy termed MiMe (Mitosis-instead-of-Meiosis, Table 2, Fig. 1A) (d’Erfurth et al. 2009), the molecular basis of which follows from functional study of each respective meiotic event.
(i) Recombination in meiosis is initiated with the formation of DNA double-strand breaks (DSB), and a series of proteins catalyzing this step are conserved from yeast to humans and plants. The SPO11 (DNA topoisomerase) complex is a tetramer composed of two copies of TOPOVIB and one copy each of SPO11-1 and SPO11-2 (Grelon et al. 2001; Hartung et al. 2007; Stacey et al. 2006; Vrielynck et al. 2016) and acts as a major player to catalyze DSB formation. In addition to the SPO11 complex, three more proteins – (Putative Recombination initiation Defect) PRD1, PRD2 and PRD3/PAIR1 – are also required for DSB formation (De Muyt et al. 2007, 2009; Nonomura et al. 2004). Mutation of any of these genes completely eliminates the recombination process. The spo11-1, prd1, pdr2 and prd3/pair1 were each shown to be efficient to generate MiMe (d’Erfurth et al. 2009; Mieulet et al. 2016) as would certainly any mutation that abolishes recombination (e.g., spo11-2 and topoVIb). An additional gene, DFO, is essential for formation of DSB in Arabidopsis, but its homologs have not been functionally analyzed in other plant species (Zhang et al. 2012). Additional genes, like SDS (Wu et al. 2015), PCH2/OsCRC1 (Miao et al. 2013), and P31comet (Ji et al. 2016), are involved in the initiation of meiotic recombination in rice but play a downstream role in the recombination process in Arabidopsis (Balboni et al. 2020; Lambing et al. 2015) (De Muyt et al. 2009; Wijeratne et al. 2006). Many other genes are needed at later stages of recombination (Mercier et al. 2015; Wang and Copenhaver 2018), but their mutation does not completely abolish recombination, making them not suitable for engineering apomeiosis with the MiMe concept.
(ii) Monopolar orientation of sister chromatids in meiosis is the second key difference between meiosis and mitosis. In mitosis, when two newly synthesized sister chromatids align at metaphase, their kinetochore (a protein complex at centromeres where the spindle binds) orients toward opposite poles, and as a result, sister chromatids move to the opposite poles. The opposite orientation of the sister kinetochores is called bipolar orientation. In contrast to mitosis, in meiosis each homologous chromosome consists of two sister chromatids that co-orient toward the same pole, a scenario known as monopolar orientation. The monopolar orientation at meiosis persists even in the absence of COs (e.g., spo11-1 mutants; Grelon et al. 2001), and sister chromatids segregate together. As they lack crossovers, the homologous chromosomes segregate randomly at meiosis I, leading to aneuploidy. Loss of monopolar orientation of the kinetochores, together with the complete abolishment of COs, allows equational segregation of chromatids at the first meiotic division, mimicking mitosis (d’Erfurth et al. 2009; Mieulet et al. 2016). In plants, the monopolar orientation of sister kinetochores at metaphase I relies on the cohesin complex (Chelysheva et al. 2005). Cohesin is a four-protein complex that forms a ring-like structure and keeps sister chromatids together after replication (Anderson et al. 2002; Gruber et al. 2003; Haering et al. 2002; Nasmyth 2002). In plants, one cohesion subunit has a meiosis-specific variant, REC8 (Bai et al. 1999; Bhatt et al. 1999). Combining the rec8 mutation with abolition of recombination results in a mitotic-like division at meiosis I with separation of the sister chromatids (Fig. 1A). However, the meiocyte then undergoes a second round of division and free chromatids segregate randomly, causing sterility (Chelysheva et al. 2005; d’Erfurth et al. 2009). Mutations of the other cohesin subunits are embryonic-lethal (Lam et al. 2005; Liu et al. 2002; Tzafrir et al. 2002) and thus cannot be used to generate apomeiosis. The cohesin subunit SCC3 has been shown to be crucial for monopolar orientation, as a weak mutant allele of SCC3 induces loss of monopolar orientation at meiosis I, but also displays somatic developmental defects (Chelysheva et al. 2005). Similarly, a kinetochore protein, MIS12, is essential in plants, but was shown by RNAi to be involved in monopolar orientation in maize (Li and Dawe 2009; Sato et al. 2005). Some meiosis-specific proteins, such as Mam1 in Saccharomyces cerevisiae, Moa1 in Schizosaccharomyces pombe, and Meikin in mammals enforce monopolar orientation, but equivalent proteins in plants have yet to be identified (Kim et al. 2015; Toth et al. 2000; Yokobayashi and Watanabe 2005). If such function exists in plants, they could be alternatives to REC8 to engineer apomeiosis.
(iii) Two rounds of division The two consecutive rounds of division during meiosis constitute a third key difference from mitosis. In spo11 rec8, the first division mimics a mitotic division (see above), but the second division still occurs, leading to meiotic catastrophe. A solution to this problem was presented by the identification of the Arabidopsis mutant osd1 (omission of second division1) (d’Erfurth et al. 2009). OSD1 is an APC/C (anaphase promoting complex/cyclosome) regulator, and its mutation causes skipping of meiosis II and consequently the generation of diploid spores and gametes. The frequency of diploid gametes in osd1 is 100% for males and 85% for females (d’Erfurth et al. 2009). The diploid gametes of osd1 are recombinant because recombination and homologous chromosome segregation still occur at meiosis I (d’Erfurth et al. 2009).
UVI4 is a paralog of Arabidopsis OSD1 that is also an APC/C inhibitor (Cromer et al. 2012; Heyman et al. 2011; Iwata et al. 2011; Van Leene et al. 2010). In uvi4, meiosis is normal but high rates of endoreduplication occur in somatic tissue. The two genes have some overlapping roles as double mutants are embryonic-lethal, indicating that OSD1 and UVI4 are crucial for somatic cell division (Cromer et al. 2012; Iwata et al. 2011). In brassicas, to which Arabidopsis belongs, the OSD1 and UVI4 orthologs are easily recognized as arising from a whole-genome duplication event that occurred at the root of this clade (Lloyd et al. 2014; Mieulet et al. 2016). Beyond the Brassicaceae, OSD1/UVI4 is typically represented by a single gene except in recent polyploids. In the clade of Poaceae, an ancient independent duplication produced two gene families (Mieulet et al. 2016). One member of the duplication was functionally characterized in rice to recapitulate the Arabidopsis osd1 phenotype, designated OsOSD1 (Mieulet et al. 2016). The frequency of diploid gamete formation is 100% in males and ~ 90% in females, similar to Arabidopsis (Mieulet et al. 2016). The second gene of OSD1/UVI 4 in rice has not yet been characterized, and based on Arabidopsis, it can be speculated that the double mutant would be lethal. Although OsOSD1 is phylogenetically distant from Arabidopsis OSD1 and UVI4, it seems to have acquired a meiotic role via convergent evolution. Therefore, OSD1 homologs can be considered promising candidate genes in Brassicaceae and Poaceae for skipping meiosis II and establishing MiMe. However, many plant species outside of the Brassicaceae and Poaceae families, like tomato and melon (Solanaceae or Cucurbitaceae), contain only one copy of either OSD1 or UVI4. Therefore, it is quite possible that its mutation would be lethal. As such, an alternative for OSD1 would be helpful for making MiMe work beyond Brassicaceae and Poaceae.
In Arabidopsis, a mutant called TARDY ASYNCHRONOUS MEIOSIS (TAM)/Cyclin CYCA1;2 was found that also skips meiosis II and results in diploid gametes similarly to osd1, as it is needed for entry into meiosis II (Cromer et al. 2012; d’Erfurth et al. 2010). However, the frequency of diploid gamete formation in tam1 is nearly 95% in males but only ~ 40% in females, which is weaker compared to osd1 (Cromer et al. 2012) and represents a limitation as the unreduced female egg is the main target cell for apomixis generation. Interestingly, combining osd1 and tam1 increased the frequency of diploid gametes in females up to 99%. However, males are quasi-sterile because male meiocytes are arrested at prophase I, which may pose a problem for creating apomixis with regard to endosperm formation (Cromer et al. 2012). Although the TAM1/CyclinA1 gene family is well conserved at the sequence level, it remains to be functionally characterized beyond Arabidopsis.
Another alternative to OSD1 is THREE DIVISION MUTANT1 (TDM1), although it plays an opposite role to TAM1 or OSD1; tdm1 knockout causes an aberrant third meiotic division after normal meiosis I and II (Cromer et al. 2012; Glover et al. 1998; Ross et al. 1997). TDM1 is proposed to be a meiosis-specific APC/C component that stimulates APC/C for meiotic exit immediately following meiosis II (Cifuentes et al. 2016). It has been also proposed that TDM1-containing P-bodies reduce the expression of meiotic transcripts to ease the switch of cell fates to post-meiotic gametophyte development (Cairo et al. 2022). CYCA1;2/TAM negatively regulates TDM through phosphorylation, as mutation of a phosphorylation site (threonine 16 on Arabidopsis TDM1) dominantly provokes the tam1 phenotype, i.e., the skipping of meiosis II, leading to diploid gamete production (Cifuentes et al. 2016). Therefore, a dominant mutation of TDM1 can be used to substitute osd1 or tam for generation of MiMe (Cifuentes et al. 2016). The TDM1 protein sequence, together with its consensus phosphorylation site, is conserved across angiosperms but has yet to be functionally characterized beyond Arabidopsis.
Two more mutants, Atps1 (Arabidopsis thaliana parallel spindle I) and Jason, produce diploid gametes due to the fusion of meiotic II spindles (d’Erfurth et al. 2008; De Storme and Geelen 2011). The frequency of diploid gametes transmitted to the next generation is less than 30% in both mutants and is restricted to males (Crismani et al. 2013; d’Erfurth et al. 2008; De Storme and Geelen 2011). Therefore, Jason and Atps1 appear as less viable candidates for MiMe generation.
(iv) Combining the three turns meiosis into mitosis MiMe, a highly efficient apomeiosis, was created in Arabidopsis by combining one mutant for each function: (i) spo11 to prevent recombination, (ii) rec8 to prevent monopolar orientation, and (iii) osd1 to skip meiosis II (d’Erfurth et al. 2009). MiMe-2 was also successfully introduced to Arabidopsis by replacing osd1 with tam1, but 10–15% of the female gametes were aneuploid, most likely due to the leakiness of tam1 (Cromer et al. 2012). By comparing the original MiMe to MiMe-2, it appears that OSD1 would be the first choice for generating MiMe, but it is possible that the penetrance of the MiMe phenotype may vary in different species. The rice MiMe was also created in the same way, but by using prd3/pair1 instead of spo11 (Mieulet et al. 2016). MiMe gametes are clones of maternal cells; thus, ploidy is doubled in the subsequent generations due to the fusion of diploid gametes.
Alternatives to MiMe for engineered apomeiosis
In Arabidopsis, DYAD/SWI1 is a crucial protein for meiosis that has been shown to protect the cohesin complex during meiotic prophase (Agashe et al. 2002; Mercier et al. 2001, 2003; Yang et al. 2019). Mutation of this single gene results in 50% of progeny deriving from apomeiosis, but it is a quasi-sterile mutant, limiting applied perspectives (Marimuthu et al. 2011; Ravi et al. 2008). The AMEIOTIC mutant, which is a homolog of DYAD/SWI1, was studied in rice and maize, but displayed high sterility (Che et al. 2011; Golubovskaya et al. 1997; Pawlowski et al. 2009). In maize, the nonreduction in female 4 (nrf4) mutant forms diploid female gametes, but only about 5% of them are clonal (Fox et al. 2016). Some epigenetic regulators have also been found to have a role in the control of meiosis in plants. The maize DNA methylation mutants dmt102 and dmt103 can induce unreduced gamete formation (Garcia-Aguilar et al. 2010). The Dnr4 ortholog in maize is known as AGO104; its mutation causes apomeiosis and produces diploid gametes with a 40–70% frequency in females (Singh et al. 2011). Similarly, in Arabidopsis ARGONAUTE9 (AGO9) regulates female gamete formation through SUPPRESSOR OF GENE SILENCING3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE6 (RDR6); mutation of any of these three genes leads to the formation of multiple gametic cells (Olmedo-Monfil et al. 2010). Thus, despite requiring three distinct mutations, it appears that MiMe remains the most efficient way to engineer apomeiosis.
Engineering apomixis step 2: parthenogenesis
A fertilization checkpoint before embryogenesis
While turning off key meiotic genes is sufficient to engineer apomeiosis, the doubling of ploidy with each generation means that MiMe alone cannot produce clonal progeny. Fertilization of diploid gametes is unaffected, and thus, MiMe mutants lack a crucial component of apomixis: embryogenesis without fertilization. As previously described, double fertilization is common to most plants and results in the formation of a zygote and an endosperm progenitor cell, the former developing into an embryo and the latter required for the proper development of the embryo (West and Harada 1993). A wide range of plant taxa have been shown to possess a trait known as parthenogenesis, in which embryos spontaneously form without fertilization, giving rise to haploid or diploid progeny (Bierzychudek 1985; Nygren 1954). In the quest to engineer parthenogenesis, great strides have been made in recent decades in elucidating the genetics controlling the transition from an unfertilized ovule to a developing embryo (Table 3). Numerous studies have implicated single dominant loci in the control of parthenogenesis in gametophytic apomicts and have demonstrated that the formation of a diploid egg cell (apospory) and the formation of a diploid embryo (parthenogenesis) are controlled separately (Albertini et al. 2001; Ogawa et al. 2013; van Dijk et al. 1999). A single dominant gene, PsASGR-BBML (P. squamulatum apospory-specific genomic region BABY BOOM-like), was found to segregate with the occurrence of apospory in the grass species Pennisetum and was shown to generate diploid offspring in sexual pearl millet tetraploids (Conner et al. 2015). Transgenic lines in rice and maize carrying PsASGR-BBML using either a native P. squamulatum promoter or a DD45 promoter conferring egg cell-specific expression saw high rates of haploid embryo formation and haploid plant recovery (Conner et al. 2017), supporting the transferability and potential application of BBM-like genes in monocot crops, although whether specific expression and/or a specific function of the protein was required for haploid induction remained unclear. BABY BOOM was initially identified in Brassica napus as an APELATA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) domain-containing gene preferentially expressed in developing embryos and able to induce somatic embryo structures when ectopically expressed (Boutilier et al. 2002), suggestive of a role in morphogenesis and the regulation of embryo development. Analysis of a BBM-like gene in rice, OsBBM1, found that it can similarly induce somatic embryos, but crucially is exclusively expressed in the male genome prior to fertilization (Khanday et al. 2019). Ectopic expression of wild-type OsBBM1 under the egg cell-specific DD45 promoter induced the formation of haploid embryos, supporting a model in which expression from the male gamete during fertilization triggers BBM1 expression in the embryo (Khanday et al. 2019, Fig. 1B). These findings suggest that BBM1 acts as a male trigger for embryogenesis and shows that the fertilization checkpoint can be overridden by BBM1 misexpression in the female genome to induce parthenogenesis.
The perhaps best-described natural apomict is common dandelion (Taraxacum officinale); genetic segregation experiments from the last two decades in dandelion and its close Asteraceae relative hawkweed (Hieracium) implicate distinct loci in controlling different aspects of apomixis. In dandelion, a locus controlling diplospory (DIP) and a separate parthenogenesis (PAR) locus were identified (van Dijk et al. 1999; van Dijk and Bakx-Schotman 2004; Vijverberg et al. 2010), with a complex third component tightly linked to the PAR locus conferring autonomous endosperm (AutE) necessary for apomixis in hawkweed and dandelion (Ogawa et al. 2013; Van Dijk et al. 2020). In aposporous hawkweed, a separate loss-of-apomeiosis (LOA) locus was characterized (Catanach et al. 2006). Most recently, deletion mapping on the basis of a loss-of-parthenogenesis phenotype and clustered regularly interspaced short palindromic repeats-based mutagenesis (CRISPR-Cas9) screening of candidate genes was used to refine the PAR locus and implicate a single dominant gene in dandelion parthenogenesis (Underwood et al. 2022). The authors found that a large transposable element (miniature inverted-repeat transposable element, or MITE) inserted within the PAR gene promoter is specific to the apomictic allele and causal for parthenogenesis. The authors next transformed parthenogenesis-deficient mutants with the MITE promoter fused to a lettuce PAR homolog, finding that the MITE-containing promoter is able to restore parthenogenesis. Fusions of the dandelion-derived PAR gene with the AtEC1 egg cell promoter could similarly complement loss-of-parthenogenesis mutants in dandelion and further induce parthenogenesis in sexual lettuce, suggesting a common mechanism in both species. These findings and the presence of an EAR-repressive motif led the authors to propose that PAR may act as a repressor of an unidentified gene suppressing embryogenesis in the egg cell; the inserted transposon could allow for PAR expression within the egg cell and trigger embryogenesis independent of sperm-contributed PAR (Underwood et al. 2022, Fig. 1B). Thus, in a natural apomict, misexpression of a dominant gene in the female gametophyte can drive parthenogenesis in a similar manner to egg cell-specific expression of BBM1. While the contribution of autonomous endosperm and diplospory still appear to be required for apomixis in dandelion and await functional characterization, PAR shows immediate promise for the engineering of parthenogenesis in dicot crops.
Haploid induction to bypass fertilization
An alternative approach to inducing apomixis lies in the pairing of apomeiosis with directed genome elimination. Preventing one of the parental genomes from contributing to fertilization, often through single loss-of-function mutations, has been shown to effectively trigger haploid induction. Mutations within the centromere-specific histone CENH3 in Arabidopsis were shown to eliminate the altered parent genome contribution upon crossing to a wild-type receiver, allowing for the production of maternal or paternal haploids (Ravi and Chan 2010, Fig. 1C). Haploid induction via CENH3 has to date been successfully implemented in maize and wheat (Karimi-Ashtiyani et al. 2015; Kelliher et al. 2016; Lv et al. 2020; Wang et al. 2021), and has been further shown to improve haploid induction rates when paired with the maize Stock6 inducer (Meng et al. 2022). While haploid induction rates of up to 45% have been shown in Arabidopsis using CENH3-mediated genome elimination (Ravi and Chan 2010), induction rates have not exceeded 5–7% in maize and wheat (Lv et al. 2020; Wang et al. 2021), well below commercially used inducer lines. One advantage in both crop systems described, however, is the ability to produce both maternal and paternal haploids, and pairing with other haploid induction strategies may be effective in increasing induction rates.
An alternative mechanism that similarly relies upon the mechanism of uniparental genome elimination was identified in a loss-of-function allele of the MATRILINEAL gene (ZmMTL/PLA1/NLD). Genetic mapping by three independent research groups of maize Stock 6, a widely used mutant crucial in haploid induction, implicated a frame-shift mutation in the phospholipase ZmMTL as causal for triggering haploid induction (Gilles et al. 2017; Kelliher et al. 2017; Liu et al. 2017). ZmMTL/PLA1/NLD was found to be exclusively expressed in pollen and is likely to encode a membrane-localized phospholipase gene with pleiotropic effects on pollen development and gene expression (Gilles et al. 2017; Kelliher et al. 2017). Mutations in MTL/PLA1/NLD have to date shown promise in haploid induction in elite cultivars of the major crops maize, rice, and wheat (Kelliher et al. 2019; Liu et al. 2020; Wang et al. 2019; Yao et al. 2018; Fig. 1C). Mutation of a ZmMTL/PLA1/NLD homolog within the Phospholipase D gene class, ZmPLD3, was also shown to produce similar levels of haploids (Li et al. 2021). Single-cell sequencing revealed that high levels of aneuploidy persist in Stock-6 derived pollen when compared to post-meiotic tetrads, suggesting that the underlying basis of haploid induction in loss-of-function mtl is driven by chromosome fragmentation (Li et al. 2017). Recent studies in wheat and maize implicated reactive oxygen species (ROS) in haploid induction driven by mtl/pla1/nld (Jiang et al. 2022; Sun et al. 2022) and showed that a lipid imbalance within mtl/pla1/nld pollen results in a ROS burst that fragments paternal DNA (Jiang et al. 2022). CRISPR mutagenesis of maize genes involved in ROS production identified a peroxidase gene, ZmPOD65, that induces haploids by utilizing the same mechanism, and it was further shown that simply treating pollen with ROS-inducing agents can produce haploids in various backgrounds (Jiang et al. 2022). Whether directly inducing ROS formation may prove useful in generating haploids efficiently and in other species is not yet clear.
While MTL/PLA1/NLD appears to be present exclusively in monocots, mapping of the qhir1 locus present in the maize haploid inducer line CAU5 uncovered a novel loss-of-function allele mutation within ZmDMP, which encodes a putative DUF679 membrane protein (Zhong et al. 2019). ZmDMP was found to be preferentially expressed in pollen and to be notably conserved among both monocots and dicots. Knockout mutants generated in ZmDMP alone saw a haploid induction rate of up to 0.3% but were shown to significantly increase haploid induction in the mtl/pla1/nld background to a maximum of 7% (Zhong et al. 2019). Mutation of ZmDMP-like genes in Arabidopsis confirmed its functional conservation within dicots (Zhong et al. 2020), paving the way for successful haploid induction in legumes and brassicas (Wang et al. 2022a, b; Zhao et al. 2022). DMP-like genes thus harbor the potential of allowing higher haploid induction rates in a range of monocots and a loss-of-function strategy for engineered parthenogenesis in dicots. The mechanism of DMP-like haploid induction remains to be clarified but could provide novel means of haploid production as seen with ZmMTL/PLA1/NLD and ROS induction.
Applied apomixis
Engineered apomixis in principle and in hybrid crops
The above sections highlight significant leaps in our understanding of apomeiosis and parthenogenesis and offer means of their application, which in tandem are required to engineer apomixis in the pursuit of clonal seeds. Combining MiMe-driven apomeiosis with the described strategies to engineer parthenogenesis can produce clonal diploid gametes that develop into synthetic apomicts (Fig. 1D). Pairing of CENH3-mediated genome elimination (GEM) with MiMe to bypass both diploid embryo formation and meiotic shuffling was first described in Arabidopsis as a means to obtain clonal seeds (Marimuthu et al. 2011). Over a third of the resultant progeny of GEM and MiMe crosses were diploid and identical to the maternal parent; repeated crossing with GEM could further fix the genetic information in subsequent generations (Marimuthu et al. 2011). Despite low seed viability and the requirement for repeated crossing, this work demonstrated that synthetic clonal propagation is feasible and yields products that are identical to those of natural apomixis. Following the finding that MiMe could be extended to rice (Mieulet et al. 2016), synthetic and autonomous clonal reproduction was first demonstrated in a major monocot crop using either BBM1- or MTL-based parthenogenesis induction (Khanday et al. 2019; Wang et al. 2019). By skipping meiosis using MiMe and targeting OsBBM1 expression to the egg cell, heterozygous SNPs present in the maternal parent were maintained for more than two generations (Khanday et al. 2019). Apomictic frequencies of up to 29% were obtained; the authors reasoned that incomplete parthenogenesis rather than variation in endosperm ploidy limits this frequency, as no defects in seed size or development were observed. It was recently demonstrated that improvements in parthenogenetic induction can in fact significantly increase the frequency of clonal progeny obtained, notably in a commercial F1 hybrid (Vernet et al. 2022). This work provides further support that engineering autonomous endosperm may not be necessary for efficient synthetic apomixis. Apomictic frequencies of greater than 95% could be maintained for at least three generations when a single DD45-BBM1-MiMe cassette was employed, well above previous frequencies (Khanday et al. 2019). While the source of the increased efficiency is unclear, one hypothesis proposes that the all-in-one construct enhances expression of BBM1 and underlies the difference. Deficiencies in fertility may be attributed to incomplete penetrance of osd1 loss of function, resulting in continued production of a low level of haploid gametes following female meiosis (Mieulet et al. 2016). This is functionally distinct from parthenogenesis and the problem could potentially be addressed by using alternative regulators of meiotic progression (d’Erfurth et al. 2010). One piece of evidence suggests that egg cell-directed OsBBM1 expression can induce the development of a second embryo from egg cell-adjacent synergid cells, the consequences of which, however, are unclear (Junhao et al. 2022). The alternative strategy pairs null OsMTL alleles with MiMe in a single gene editing cassette to produce clonal progeny directly in a hybrid variety (Wang et al. 2019), although with significant reductions in fertility compared to BBM1 strategies, a known consequence of OsMTL-driven haploid induction (Yao et al. 2018). It was recently proposed that incomplete genome elimination by osmtl may be responsible for observed losses in fertility (Liu et al. 2022). Despite this limitation, the maintenance of heterosis in engineered apomict crops has been reported using both MTL and BBM1 strategies (Liu et al. 2022; Vernet et al. 2022), supplementing work in Hieracium proving that natural apomicts can indeed transmit complex phenotypes derived from hybrids (Sailer et al. 2016). Reduced seed viability remains a hurdle and appears to arise from multiple sources, but strategies that mitigate against this problem exist. The implementation of synthetic apomixis in other economically important crops has yet to be reported, though the conservation of key meiotic genes (Hyde et al. 2022; Ma et al. 2018) as well as BBM, MTL, and DMP-like genes (Chen et al. 2022; Liu et al. 2020; Wang, Xia, et al. 2022a, b) offers promise. Further worth considering is the recent clarification of the locus controlling nucellar or adventitious embryony in citrus (Wang et al. 2017), a defining feature of sporophytic apomixis. Comparative genomics identified the co-segregation of citrus polyembryony with a MITE insertion in the promoter of a candidate gene termed CitRWP, possessing a domain known to influence embryogenesis (Waki et al. 2011). While polyembryony is observed in natural gametophytic apomicts, further functional validation is required and the persistence of both sexual and asexual embryos may pose problems for efficient engineered apomixis. The current body of work demonstrates that high levels of clonal progeny in crops can be obtained by sidestepping meiosis and excluding the contribution of one of the parental genomes. The combined apomeiosis-parthenogenesis provides a strategy that is fully penetrant and able to propagate hybrid vigor, though limitations remain in terms of fertility.
Outlook for engineered apomixis and potential roadblocks
It is now clear that by fine-tuning a few key regulators of meiosis and embryogenesis, one can engineer apomixis within several major crops. While exciting, the acceptance of such technologies face significant hurdles with regard to the end consumer and the legislation that regulates their use (Batalha et al. 2021; Turnbull et al. 2021). The methodologies presented in this review are termed and regulated as two separate technologies in many countries, namely genetic engineering or modification (GM) and genome editing (GE). While the former is subject to extensive regulation by many governing bodies, the latter has been granted exemption by some governments (S. M. Schmidt et al. 2020), acknowledging the fact that GE does not introduce large pieces of foreign DNA and is thus similar in outcome to the product of conventional and mutagenesis breeding (Turnbull et al. 2021). This discrepancy has had a profound influence on the commercialization of CRISPR-based genome-edited products (Martin-Laffon et al. 2019). Despite continued review of such policies (Friedrichs et al. 2019) and proposals to integrate GE technologies (Huang et al. 2016), a mismatch nonetheless exists between the pace of acceptance and the acceleration of their development. Utilization of MiMe and MTL mutations to engineer apomeiosis and parthenogenesis may fall in the category of GE, as they require simple edits through CRISPR-based approaches. Misexpression of BBM1/PAR in the developing embryo, however, is dependent upon the stable introduction of recombinant DNA, namely alternative promoter sequences, and thus has the potential to see greater regulatory pushback regardless of whether these sequences remain plant-derived.
A number of strategies have been reported in recent years that aim to assuage some legislative and technical concerns related to genome editing. While the product of CRISPR-mediated genome editing does not retain large pieces of inserted foreign DNA, introducing edits often requires stable expression of a CRISPR/Cas9 transgene cassette through transformation. Genetic segregation can remove the transgene in subsequent generations, but this approach would not be possible in a clonally reproducing synthetic apomict. One tactic couples the transformation of a CRISPR/Cas9 cassette with the expression of two genes that trigger cell death during embryo development and within the male gametophyte, termed TKC (Transgene Killer CRISPR) (He et al. 2018). Expression of the toxic BARNASE gene during rice embryo development and rice CMS2 conferring male sterility, together with a genome editing construct, can induce cell death in reproductive tissue containing transgenes. This approach ensures that any seeds containing the transformed construct do not develop and precludes the need for transgene removal by segregation. Alternatively, a method that does not require insertion of foreign DNA introduces CRISPR/Cas9-bundled ribonucleoproteins (RNPs) to direct editing. Several reports have demonstrated success in delivering RNPs into protoplasts, embryos, or zygotes of crop species to confer entirely DNA-free genome editing (Liang et al. 2017; Toda et al. 2019; Woo et al. 2015). Both TKC and RNP editing methods have the added benefit of reducing possible off-targets during editing, a concern common to both regulators and researchers (Hahn and Nekrasov 2019; S. M. Schmidt et al. 2020). Recent work also demonstrated that CRISPR cassettes can be virally delivered to induce heritable mutations in a model system (Ma et al. 2020), opening the door to further transgene-free options. While the above represent methods to resolve potential GE concerns, incorporation of stable transgenes via GM is still necessary in several strategies to engineer apomixis and remains a possible roadblock for product development.
Following its acceptance by regulators, questions still remain as to how exactly engineered apomixis can fit into modern breeding schemes. Apomicts derived from dominant mutations have been suggested to act as pollen donors for crossing to sexual elite varieties (van Dijk et al. 2016), a strategy that could greatly reduce the time and resource-intensive breeding cycles and one that remains compatible with synthetic apomixis strategies described above. Alternatively, direct manipulation of elite varieties could further shorten and simplify the breeding cycle, provided that transformation remains feasible. An additional concern lies in the intellectual property rights surrounding apomixis technology; while frameworks are currently limited, a proposal put forth by apomixis researchers in 1998 stressed the importance of maintaining equitable access to such technologies for the benefit of global food security (Grossniklaus et al. 1998). Lastly, the emergence of synthetic apomicts will undoubtedly influence the economics of seed production by allowing hybrids to self-propagate similar to current elite non-hybrid pure lines. In the context of a seed market that is evenly split between public institutions, private industry, and farmer seed-saving, it could have a net positive effect for each group. It has been proposed that savings to both growers and industries could reach upwards of billions of dollars per year (Spillane et al. 2004), while simultaneously facilitating true seed production, diversifying hybrid breeding, and consequently improving adaptability in changing climates.
References
Agashe B, Prasad CK, Siddiqi I (2002) Identification and analysis of DYAD: a gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129(16):3935–3943. https://doi.org/10.1242/dev.129.16.3935
Albertini E, Porceddu A, Ferranti F, Reale L, Barcaccia G, Romano B, Falcinelli M (2001) Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sex Plant Reprod 14(4):213–217. https://doi.org/10.1007/s00497-001-0116-2
Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156(3):419–424. https://doi.org/10.1083/jcb.200111002
Asker SE, Jerling L (1992) Apomixis in plants. CRC Press, London
Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11(3):417–430. https://doi.org/10.1105/tpc.11.3.417
Balboni M, Yang C, Komaki S, Brun J, Schnittger A (2020) COMET functions as a PCH2 cofactor in regulating the HORMA domain protein ASY1. Curr Biol 30(21):4113-4127.e6. https://doi.org/10.1016/j.cub.2020.07.089
Barke BH, Karbstein K, Daubert M, Hörandl E (2020) The relation of meiotic behaviour to hybridity, polyploidy and apomixis in the Ranunculus auricomus complex (Ranunculaceae). BMC Plant Biol 20(1):523. https://doi.org/10.1186/s12870-020-02654-3
Batalha L, Foroni F, Jones BJ (2021) All plant breeding technologies are equal, but some are more equal than others: the case of GM and mutagenesis. Frontiers in Plant Science 12:258
Benyahya F, Nadaud I, Da Ines O, Rimbert H, White C, Sourdille P (2020) SPO11.2 is essential for programmed double-strand break formation during meiosis in bread wheat (Triticum aestivum L.). Plant J 104(1):30–43
Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19(4):463–472. https://doi.org/10.1046/j.1365-313X.1999.00548.x
Bierzychudek P (1985) Patterns in plant parthenogenesis. Experientia 41(10):1255–1264. https://doi.org/10.1007/BF01952068
Böcher T (1952) Cytological and embryological studies in the Amphiapomictic Arabis Holboellii. Complex 5(1):57–57
Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA, Custers JBM, Campagne MMV (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14(8):1737–1749. https://doi.org/10.1105/tpc.001941
Brozova V, Koutecky P, Dolezal J (2019) Plant apomixis is rare in Himalayan high-alpine flora. Sci Rep 9:14386. https://doi.org/10.1038/s41598-019-50907-5
Burson BL (1997) Apomixis and sexuality in some paspalum species. Crop Sci 37(4):555
Caetano APS, Cortez PA, Teixeira SP, Oliveira PE, Carmello-Guerreiro SM (2018) Unusual diversity of apomictic mechanisms in a species of Miconia, Melastomataceae. Plant Syst Evol 304(3):343–355. https://doi.org/10.1007/s00606-017-1480-1
Caetano APS, Oliveira PE (2022) Apomixis in Melastomataceae. In: Goldenberg R, Michelangeli FA, Almeda F (eds) Systematics, evolution, and ecology of Melastomataceae. Springer, Berlin, pp 563–583
Cairo A, Vargova A, Shukla N, Capitao C, Mikulkova P, Valuchova S, Pecinkova J, Bulankova P, Riha K (2022) Meiotic exit in Arabidopsis is driven by P-body–mediated inhibition of translation. Science 377(6606):629–634. https://doi.org/10.1126/science.abo0904
Carman JG (1997) Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol J Lin Soc 61(1):51–94. https://doi.org/10.1111/j.1095-8312.1997.tb01778.x
Catanach AS, Erasmuson SK, Podivinsky E, Jordan BR, Bicknell R (2006) Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc Natl Acad Sci 103(49):18650–18655. https://doi.org/10.1073/pnas.0605588103
Che L, Tang D, Wang K, Wang M, Zhu K, Yu H, Gu M, Cheng Z (2011) OsAM1 is required for leptotene-zygotene transition in rice. Cell Res 21(4):5558
Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mézard C, Grelon M (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118(20):4621–4632. https://doi.org/10.1242/jcs.02583
Chen B, Maas L, Figueiredo D, Zhong Y, Reis R, Li M, Horstman A, Riksen T, Weemen M, Liu H, Siemons C, Chen S, Angenent GC, Boutilier K (2022) BABY BOOM regulates early embryo and endosperm development. Proc Natl Acad Sci 119(25):e2201761119. https://doi.org/10.1073/pnas.2201761119
Chen SH, Guja LK, Schmidt-Lebuhn AN (2019) Conservation implications of widespread polyploidy and apomixis: a case study in the genus Pomaderris (Rhamnaceae). Conserv Genet 20(4):917–926. https://doi.org/10.1007/s10592-019-01184-2
Cifuentes M, Jolivet S, Cromer L, Harashima H, Bulankova P, Renne C, Crismani W, Nomura Y, Nakagami H, Sugimoto K, Schnittger A, Riha K, Mercier R (2016) TDM1 regulation determines the number of meiotic divisions. PLOS Genet 12(2):e1005856. https://doi.org/10.1371/journal.pgen.1005856
Conner JA, Mookkan M, Huo H, Chae K, Ozias-Akins P (2015) A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci USA 112(36):11205–11210. https://doi.org/10.1073/pnas.1505856112
Conner JA, Ozias-Akins P (2017) Apomixis: engineering the ability to harness hybrid vigor in crop plants. In: Schmidt A (ed) Plant germline development: methods and protocols. Springer, Berlin, pp 17–34. https://doi.org/10.1007/978-1-4939-7286-9_2
Conner JA, Podio M, Ozias-Akins P (2017) Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod 30(1):41–52. https://doi.org/10.1007/s00497-017-0298-x
Cosendai A-C, Hörandl E (2010) Cytotype stability, facultative apomixis and geographical parthenogenesis in Ranunculus kuepferi (Ranunculaceae). Ann Bot 105(3):457–470. https://doi.org/10.1093/aob/mcp304
Crismani W, Girard C, Mercier R (2013) Tinkering with meiosis. J Exp Bot 64(1):55–65. https://doi.org/10.1093/jxb/ers314
Cromer L, Heyman J, Touati S, Harashima H, Araou E, Girard C, Horlow C, Wassmann K, Schnittger A, Veylder LD, Mercier R (2012) OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLOS Genet 8(7):e1002865. https://doi.org/10.1371/journal.pgen.1002865
d’Erfurth I, Cromer L, Jolivet S, Girard C, Horlow C, Sun Y, To JPC, Berchowitz LE, Copenhaver GP, Mercier R (2010) The CYCLIN-A CYCA1;2/TAM is required for the Meiosis I to Meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLOS Genet 6(6):e1000989. https://doi.org/10.1371/journal.pgen.1000989
d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R (2009) Turning Meiosis into Mitosis. PLOS Biol 7(6):e1000124. https://doi.org/10.1371/journal.pbio.1000124
d’Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Simon M, Jenczewski E, Mercier R (2008) Mutations in AtPS1 (Arabidopsis thaliana Parallel Spindle 1) lead to the production of diploid pollen grains. PLOS Genet 4(11):e1000274. https://doi.org/10.1371/journal.pgen.1000274
De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, Lainé-Choinard S, Pelletier G, Mercier R, Nogué F, Grelon M (2009) A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLOS Genet 5(9):e1000654. https://doi.org/10.1371/journal.pgen.1000654
De Muyt A, Vezon D, Gendrot G, Gallois J-L, Stevens R, Grelon M (2007) AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J 26(18):4126–4137. https://doi.org/10.1038/sj.emboj.7601815
De Storme N, Geelen D (2011) The Arabidopsis Mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in Meiosis II. Plant Physiol 155(3):1403–1415. https://doi.org/10.1104/pp.110.170415
Dickinson TA (2018) Sex and Rosaceae apomicts. TAXON 67(6):1093–1107
Dobes C, Mitchell-Olds T, Koch MA (2004) Intraspecific diversification in North American Boechera stricta (= Arabis drummondii), Boechera xdivaricarpa, and Boechera holboellii (Brassicaceae) inferred from nuclear and chloroplast molecular markers—an integrative approach. Am J Bot 91(12):2087–2101. https://doi.org/10.3732/ajb.91.12.2087
Dobeš Ch, Milosevic A, Prohaska D, Scheffknecht S, Sharbel TF, Hülber K (2013) Reproductive differentiation into sexual and apomictic polyploid cytotypes in Potentilla puberula (Potentilleae, Rosaceae). Ann Bot 112(6):1159–1168. https://doi.org/10.1093/aob/mct167
Ernst A (1909) Apogamie bei Burmannia coelestis. Berichte Der Deutschen Botanischen Gesellschaft 27:157–168
Fayos I, Meunier AC, Vernet A, Navarro-Sanz S, Portefaix M, Lartaud M, Bastianelli G, Perin C, Nicolas A, Guiderdoni E (2020) Assessment of the roles of SPO11-2 and SPO11-4 in meiosis in rice using CRISPR/Cas9 mutagenesis. J Exp Bot 71(22):7046–7058. https://doi.org/10.1093/jxb/eraa391
Firetti F (2017) Apomixis in neotropical vegetation. In: Vegetation. IntechOpen. https://doi.org/10.5772/intechopen.71856
Firetti-Leggieri F, Lohmann LG, Alcantara S, da Costa IR, Semir J (2013) Polyploidy and polyembryony in Anemopaegma (Bignonieae, Bignoniaceae). Plant Reprod 26(1):43–53. https://doi.org/10.1007/s00497-012-0206-3
Fox TW, Albertsen MC, Williams ME, Lawit SJ, Chamberlin MA, Grossniklaus U, Brunner GA, Chumak N, De Asis JB, Pasquer F (2016) Methods and compositions for the production of unreduced, non-recombined gametes and clonal offspring (Patent No. WO 2016/179522 A1). https://lens.org/010-079-811-462-856
Friedrichs S, Takasu Y, Kearns P, Dagallier B, Oshima R, Schofield J, Moreddu C (2019) An overview of regulatory approaches to genome editing in agriculture. Biotechnol Res Innov 3(2):208–220. https://doi.org/10.1016/j.biori.2019.07.001
Fu L-F, Su L-Y, Mallik A, Wen F, Wei Y-G (2017) Cytology and sexuality of 11 species of Elatostema (Urticaceae) in limestone karsts suggests that apomixis is a recurring phenomenon. Nord J Bot 35(2):251–256. https://doi.org/10.1111/njb.01281
Galla G, Vogel H, Sharbel TF, Barcaccia G (2015) De novo sequencing of the Hypericum perforatum L. flower transcriptome to identify potential genes that are related to plant reproduction sensu lato. BMC Genom 16(1):254. https://doi.org/10.1186/s12864-015-1439-y
Garcia-Aguilar M, Michaud C, Leblanc O, Grimanelli D (2010) Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22(10):3249–3267. https://doi.org/10.1105/tpc.109.072181
Gilles LM, Khaled A, Laffaire J-B, Chaignon S, Gendrot G, Laplaige J, Berges H, Beydon G, Bayle V, Barret P, Comadran J, Martinant J-P, Rogowsky PM, Widiez T (2017) Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. Embo J 36(6):707–717. https://doi.org/10.15252/embj.201796603
Glover J, Grelon M, Craig S, Chaudhury A, Dennis E (1998) Cloning and characterization of MS5 from Arabidopsis: a gene critical in male meiosis. Plant J 15(3):345–356. https://doi.org/10.1046/j.1365-313X.1998.00216.x
Golubovskaya I, Avalkina N, Sheridan WF (1997) New insights into the role of the maize ameiotic1 locus. Genetics 147(3):1339–1350. https://doi.org/10.1093/genetics/147.3.1339
Golubovskaya IN, Hamant O, Timofejeva L, Wang C-JR, Braun D, Meeley R, Cande WZ (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119(16):3306–3315. https://doi.org/10.1242/jcs.03054
Gonzalez AM, Sato HA, Marazzi B (2019) Embryology in Helosis cayennensis (Balanophoraceae): structure of female flowers, fruit, endosperm and embryo. Plants 8(3):3. https://doi.org/10.3390/plants8030074
Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20(3):589–600. https://doi.org/10.1093/emboj/20.3.589
Grossniklaus U, Koltunow A, van Campagne M, L (1998) A bright future for apomixis. Trends Plant Sci 3(11):415–416. https://doi.org/10.1016/S1360-1385(98)01338-7
Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112(6):765–777. https://doi.org/10.1016/S0092-8674(03)00162-4
Haering CH, Löwe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9(4):773–788. https://doi.org/10.1016/S1097-2765(02)00515-4
Hafidh S, Honys D (2021) Reproduction multitasking: the male gametophyte. Annu Rev Plant Biol 72:581–614. https://doi.org/10.1146/annurev-arplant-080620-021907
Hahn F, Nekrasov V (2019) CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep 38(4):437–441. https://doi.org/10.1007/s00299-018-2355-9
Hand ML, Koltunow AMG (2014) The genetic control of apomixis: asexual seed formation. Genetics 197(2):441–450. https://doi.org/10.1534/genetics.114.163105
Hartung F, Wurz-Wildersinn R, Fuchs J, Schubert I, Suer S, Puchta H (2007) The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19(10):3090–3099. https://doi.org/10.1105/tpc.107.054817
He Y, Zhu M, Wang L, Wu J, Wang Q, Wang R, Zhao Y (2018) Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol Plant 11(9):1210–1213. https://doi.org/10.1016/j.molp.2018.05.005
Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Kamei CLA, De Jaeger G, Koncz C, De Veylder L (2011) Arabidopsis ultraviolet-B-insensitive4 maintains cell division activity by temporal Inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23(12):4394–4410. https://doi.org/10.1105/tpc.111.091793
Hochholdinger F, Baldauf JA (2018) Heterosis in plants. Curr Biol 28(18):R1089–R1092. https://doi.org/10.1016/j.cub.2018.06.041
Hojsgaard D, Hörandl E (2019) The rise of apomixis in natural plant populations. Front Plant Sci 10:55. https://doi.org/10.3389/fpls.2019.00358
Hojsgaard D, Klatt S, Baier R, Carman JG, Hörandl E (2014) Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. Crit Rev Plant Sci 33(5):414–427. https://doi.org/10.1080/07352689.2014.898488
Huang S, Weigel D, Beachy RN, Li J (2016) A proposed regulatory framework for genome-edited crops. Nat Genet 48(2):2. https://doi.org/10.1038/ng.3484
Hyde L, Osman K, Winfield M, Sanchez-Moran E, Higgins JD, Henderson IR, Sparks C, Franklin FCH, Edwards KJ (2022) Identification, characterization, and rescue of CRISPR/Cas9 generated wheat SPO11-1 mutants. Plant Biotechnol J. https://doi.org/10.1111/pbi.13961
Iwata E, Ikeda S, Matsunaga S, Kurata M, Yoshioka Y, Criqui M-C, Genschik P, Ito M (2011) GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in arabidopsis. Plant Cell 23(12):4382–4393. https://doi.org/10.1105/tpc.111.092049
Ji J, Tang D, Shen Y, Xue Z, Wang H, Shi W, Zhang C, Du G, Li Y, Cheng Z (2016) P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proc Natl Acad Sci 113(38):10577–10582. https://doi.org/10.1073/pnas.1607334113
Jiang C, Sun J, Li R, Yan S, Chen W, Guo L, Qin G, Wang P, Luo C, Huang W, Zhang Q, Fernie AR, Jackson D, Li X, Yan J (2022) A reactive oxygen species burst causes haploid induction in maize. Mol Plant 15(6):943–955. https://doi.org/10.1016/j.molp.2022.04.001
Junhao D, Xia Y, Zhan Y, Zhang L, Tang N, Wang Y, Tian J, Deng H, Cao M (2022) High-frequency of twin-embryo seeds with clonal and sexually produced embryos drives apomixis in hybrid ric. https://doi.org/10.21203/rs.3.rs-2009585/v1
Karimi-Ashtiyani R, Ishii T, Niessen M, Stein N, Heckmann S, Gurushidze M, Banaei-Moghaddam AM, Fuchs J, Schubert V, Koch K, Weiss O, Demidov D, Schmidt K, Kumlehn J, Houben A (2015) Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc Natl Acad Sci USA 112(36):11211–11216. https://doi.org/10.1073/pnas.1504333112
Kelliher T, Starr D, Richbourg L, Chintamanani S, Delzer B, Nuccio ML, Green J, Chen Z, McCuiston J, Wang W, Liebler T, Bullock P, Martin B (2017) MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542(7639):105. https://doi.org/10.1038/nature20827
Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, McCuiston J, Gu W, Sun Y, Strebe T, Roberts J, Bate NJ, Que Q (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37(3):3. https://doi.org/10.1038/s41587-019-0038-x
Kelliher T, Starr D, Wang W, McCuiston J, Zhong H, Nuccio ML, Martin B (2016) Maternal haploids are preferentially induced by CENH3-tailswap transgenic complementation in maize. Front Plant Sci 7:414. https://doi.org/10.3389/fpls.2016.00414
Kellogg EA (1990) Variation and species limits in agamospermous grasses. Syst Bot 15(1):112–123. https://doi.org/10.2307/2419021
Khanday I, Skinner D, Yang B, Mercier R, Sundaresan V (2019) A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565(7737):91. https://doi.org/10.1038/s41586-018-0785-8
Kim J, Ishiguro K, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas AM, Takeda N, Sakakibara Y, Kitajima TS, Tanno Y, Sakuno T, Watanabe Y (2015) Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517(7535):7535. https://doi.org/10.1038/nature14097
Koltunow AMG (1994) Apomixis—other pathways for reproductive development in angiosperms. In: Williams EG, Clarke AE, Knox RB (eds) Genetic control of self-incompatibility and reproductive development in flowering plants. Springer, Berlin, pp 486–512. https://doi.org/10.1007/978-94-017-1669-7_23
Koltunow AMG, Grossniklaus U (2003) Apomixis: a developmental perspective. Annu Rev Plant Biol 54:547–574. https://doi.org/10.1146/annurev.arplant.54.110901.160842
Koltunow AMG, Johnson SD, Rodrigues JCM, Okada T, Hu Y, Tsuchiya T, Wilson S, Fletcher P, Ito K, Suzuki G, Mukai Y, Fehrer J, Bicknell RA (2011) Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella, in which two dominant loci function to enable apomixis. Plant J 66(5):890–902. https://doi.org/10.1111/j.1365-313X.2011.04556.x
Kuppu S, Ron M, Marimuthu MPA, Li G, Huddleson A, Siddeek MH, Terry J, Buchner R, Shabek N, Comai L, Britt AB (2020) A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol J 18(10):2068–2080. https://doi.org/10.1111/pbi.13365
Kuppu S, Tan EH, Nguyen H, Rodgers A, Comai L, Chan SWL, Britt AB (2015) Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLOS Genet 11(9):e1005494. https://doi.org/10.1371/journal.pgen.1005494
Lam WS, Yang X, Makaroff CA (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci 118(14):3037–3048. https://doi.org/10.1242/jcs.02443
Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, Yang J, Armstrong SJ, Mechtler K, Roitinger E, Franklin FCH (2015) Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLOS Genet 11(7):51005372. https://doi.org/10.1371/journal.pgen.1005372
Li X, Dawe RK (2009) Fused sister kinetochores initiate the reductional division in meiosis I. Nat Cell Biol 11(9):9. https://doi.org/10.1038/ncb1923
Li X, Meng D, Chen S, Luo H, Zhang Q, Jin W, Yan J (2017) Single nucleus sequencing reveals spermatid chromosome fragmentation as a possible cause of maize haploid induction. Nat Commun 8:991. https://doi.org/10.1038/s41467-017-00969-8
Li Y, Lin Z, Yue Y, Zhao H, Fei X, Lizhu E, Liu C, Chen S, Lai J, Song W (2021) Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat Plants 7(12):1579. https://doi.org/10.1038/s41477-021-01037-2
Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8(1):1. https://doi.org/10.1038/ncomms14261
Liu C, He Z, Zhang Y, Hu F, Li M, Liu Q, Huang Y, Wang J, Zhang W, Wang C, Wang K (2022) Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice. Plant Commun. https://doi.org/10.1016/j.xplc.2022.100470
Liu C, Li X, Meng D, Zhong Y, Chen C, Dong X, Xu X, Chen B, Li W, Li L, Tian X, Zhao H, Song W, Luo H, Zhang Q, Lai J, Jin W, Yan J, Chen S (2017) A 4-bp insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize. Mol Plant 10(3):520–522. https://doi.org/10.1016/j.molp.2017.01.011
Liu C, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AAM, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J Cell Mol Biol 29(4):405–415. https://doi.org/10.1046/j.1365-313x.2002.01224.x
Liu H, Wang K, Jia Z, Gong Q, Lin Z, Du L, Pei X, Ye X (2020) Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J Exp Bot 71(4):1337–1349. https://doi.org/10.1093/jxb/erz529
Lloyd AH, Ranoux M, Vautrin S, Glover N, Fourment J, Charif D, Choulet F, Lassalle G, Marande W, Tran J, Granier F, Pingault L, Remay A, Marquis C, Belcram H, Chalhoub B, Feuillet C, Bergès H, Sourdille P, Jenczewski E (2014) Meiotic gene evolution: can you teach a new dog new tricks? Mol Biol Evol 31(7):1724–1727. https://doi.org/10.1093/molbev/msu119
Lv J, Yu K, Wei J, Gui H, Liu C, Liang D, Wang Y, Zhou H, Carlin R, Rich R, Lu T, Que Q, Wang WC, Zhang X, Kelliher T (2020) Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat Biotechnol 38(12):514. https://doi.org/10.1038/s41587-020-0728-4
Ma G, Zhang W, Liu L, Chao WS, Gu YQ, Qi L, Xu SS, Cai X (2018) Cloning and characterization of the homoeologous genes for the Rec8-like meiotic cohesin in polyploid wheat. BMC Plant Biol 18:224. https://doi.org/10.1186/s12870-018-1442-y
Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants 6(7):7. https://doi.org/10.1038/s41477-020-0704-5
Majeský L, Krahulec F, Vašut RJ (2017) How apomictic taxa are treated in current taxonomy: a review. TAXON 66(5):1017–1040
Marimuthu MPA, Jolivet S, Ravi M, Pereira L, Davda JN, Cromer L, Wang L, Nogué F, Chan SWL, Siddiqi I, Mercier R (2011) Synthetic clonal reproduction through seeds. Science 331(6019):876–876. https://doi.org/10.1126/science.1199682
Martin-Laffon J, Kuntz M, Ricroch AE (2019) Worldwide CRISPR patent landscape shows strong geographical biases. Nat Biotechnol 37(6):6. https://doi.org/10.1038/s41587-019-0138-7
Meng D, Luo H, Dong Z, Huang W, Liu F, Li F, Chen S, Yu H, Jin W (2022) Overexpression of modified CENH3 in Maize Stock6-derived inducer lines can effectively improve maternal haploid induction rates. Front Plant Sci 13:5110. https://doi.org/10.3389/fpls.2022.892055
Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FCH (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130(14):3309–3318
Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M (2015) The molecular biology of meiosis in plants. Annu Rev Plant Biol 66(1):297–327. https://doi.org/10.1146/annurev-arplant-050213-035923
Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefèvre F, Pelletier G, Horlow C (2001) SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15(14):1859–1871. https://doi.org/10.1101/gad.203201
Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z (2013) Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 25(8):2998–3009. https://doi.org/10.1105/tpc.113.113175
Mieulet D, Jolivet S, Rivard M, Cromer L, Vernet A, Mayonove P, Pereira L, Droc G, Courtois B, Guiderdoni E, Mercier R (2016) Turning rice meiosis into mitosis. Cell Res 26(11):1242–1254. https://doi.org/10.1038/cr.2016.117
Mráz P, Zdvořák P (2019) Reproductive pathways in Hieracium s.s. (Asteraceae): strict sexuality in diploids and apomixis in polyploids. Ann Bot 123(2):391–403. https://doi.org/10.1093/aob/mcy137
Nakano M, Kigoshi K, Shimizu T, Endo T, Shimada T, Fujii H, Omura M (2013) Characterization of genes associated with polyembryony and in vitro somatic embryogenesis in Citrus. Tree Genet Genomes 9(3):795–803. https://doi.org/10.1007/s11295-013-0598-8
Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 297(5581):559–565. https://doi.org/10.1126/science.1074757
Nogler GA (1984) Gametophytic apomixis. In: Johri BM (ed) Embryology of Angiosperms. Springer, Berlin, pp 475–518. https://doi.org/10.1007/978-3-642-69302-1_10
Nonomura K-I, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N (2004) The novel gene homologous pairing aberration in rice meiosis1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16(4):1008–1020. https://doi.org/10.1105/tpc.020701
Novak SJ, Mack RN (2000) Clonal diversity within and among introduced populations of the apomictic vine Bryonia alba (Cucurbitaceae). Can J Bot 78(11):1469–1481. https://doi.org/10.1139/b00-116
Noyes RD (2007) Apomixis in the Asteraceae: Diamonds in the rough. Funct Plant Sci Biotechnol 1(2):207–222
Nygren A (1954) Apomixis in the angiosperms. II. Botanical Rev 20(10):577–649. https://doi.org/10.1007/BF02958805
Ogawa D, Johnson SD, Henderson ST, Koltunow AMG (2013) Genetic separation of autonomous endosperm formation (AutE) from the two other components of apomixis in Hieracium. Plant Reprod 26(2):113–123. https://doi.org/10.1007/s00497-013-0214-y
Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada J-P (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464(7288):7288. https://doi.org/10.1038/nature08828
Ortiz JPA, Quarin CL, Pessino SC, Acuña C, Martínez EJ, Espinoza F, Hojsgaard DH, Sartor ME, Cáceres ME, Pupilli F (2013) Harnessing apomictic reproduction in grasses: What we have learned from Paspalum. Ann Bot 112(5):767–787. https://doi.org/10.1093/aob/mct152
Pangsuban S, Bamroongrugsa N, Kanchanapoom K, Nualsri C (2009) Facultative Apomixis in Garcinia atroviridis (Clusiaceae) and effects of different pollination regimes on reproductive success. Trop Life Sci Res 20(2):89–108
Pawlowski WP, Wang C-JR, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ (2009) Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc Natl Acad Sci 106(9):3603–3608. https://doi.org/10.1073/pnas.0810115106
Qing-Wen Z, Dian-Xiang Z, Ze-Zheng GAO, Fu-Wu X (2003) Facultative apomixis in an endangered dioecious species, Woonyoungia septentrionalis (Magnoliaceae). J Integr Plant Biol 45(11):1270
Ravi M, Chan SWL (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464(7288):7288. https://doi.org/10.1038/nature08842
Ravi M, Marimuthu MPA, Siddiqi I (2008) Gamete formation without meiosis in Arabidopsis. Nature 451(7182):7182. https://doi.org/10.1038/nature06557
Ribeiro DN, Pan Z, Duke SO, Nandula VK, Baldwin BS, Shaw DR, Dayan FE (2014) Involvement of facultative apomixis in inheritance of EPSPS gene amplification in glyphosate-resistant Amaranthus palmeri. Planta 239(1):199–212. https://doi.org/10.1007/s00425-013-1972-3
Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FCH, Jones GH (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res 5(8):551–559. https://doi.org/10.1023/A:1018497804129
Sáez L, Rosselló JA (1996) Limonium inexpectans (Plumbaginaceae), a new apomictic species from Majorca (Balearic Islands). Anales Jardín Botánico De Madrid 1(1):289–289
Sailer C, Schmid B, Grossniklaus U (2016) Apomixis allows the transgenerational fixation of phenotypes in hybrid plants. Curr Biol 26(3):331–337. https://doi.org/10.1016/j.cub.2015.12.045
Sato H, Shibata F, Murata M (2005) Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res 13(8):827–834. https://doi.org/10.1007/s10577-005-1016-3
Schmidt SM, Belisle M, Frommer WB (2020) The evolving landscape around genome editing in agriculture. EMBO Rep 21(6):e50680
Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156(3):3. https://doi.org/10.1104/pp.111.177428
Sharbel TF, Mitchell-Olds T, Dobes C, Kantama L, de Jong H (2005) Biogeographic distribution of polyploidy and B chromosomes in the apomictic Boechera holboellii complex. Cytogenet Genome Res 109(1–3):283–292. https://doi.org/10.1159/000082411
Shi W, Ji J, Xue Z, Zhang F, Miao Y, Yang H, Tang D, Du G, Li Y, Shen Y, Cheng Z (2021) PRD1, a homologous recombination initiation factor, is involved in spindle assembly in rice meiosis. New Phytol 230(2):585–600. https://doi.org/10.1111/nph.17178
Shimada T, Endo T, Fujii H, Nakano M, Sugiyama A, Daido G, Ohta S, Yoshioka T, Omura M (2018) MITE insertion-dependent expression of CitRKD1 with a RWP-RK domain regulates somatic embryogenesis in citrus nucellar tissues. BMC Plant Biol 18(1):166. https://doi.org/10.1186/s12870-018-1369-3
Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D (2011) Production of Viable Gametes without Meiosis in Maize Deficient for an ARGONAUTE Protein. Plant Cell 23(2):443–458. https://doi.org/10.1105/tpc.110.079020
Skinner DJ, Sundaresan V (2018) Recent advances in understanding female gametophyte development. F1000Research 7:804. https://doi.org/10.12688/f1000research.14508.1
Souza-Pérez M, Speroni G (2017) New apomictic pathway in Myrtaceae inferred from Psidium cattleyanum female gametophyte ontogeny. Flora 234:34–40. https://doi.org/10.1016/j.flora.2017.06.010
Spillane C, Curtis MD, Grossniklaus U (2004) Apomixis technology development—Virgin births in farmers’ fields? Nat Biotechnol 22(6):687–691. https://doi.org/10.1038/nbt976
Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K (2006) Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J 48(2):206–216. https://doi.org/10.1111/j.1365-313X.2006.02867.x
Sun G, Geng S, Zhang H, Jia M, Wang Z, Deng Z, Tao S, Liao R, Wang F, Kong X, Fu M, Liu S, Li A, Mao L (2022) Matrilineal empowers wheat pollen with haploid induction potency by triggering postmitosis reactive oxygen species activity. New Phytol 233(6):2405–2414. https://doi.org/10.1111/nph.17963
Talent N, Dickinson TA (2007) Endosperm formation in aposporous Crataegus (Rosaceae, Spiraeoideae, tribe Pyreae): Parallels to Ranunculaceae and Poaceae. New Phytol 173(2):231–249. https://doi.org/10.1111/j.1469-8137.2006.01918.x
Teppner H (1996) Adventitious embryony inNigritella (Orchidaceae). Folia Geobot 31(3):323–331. https://doi.org/10.1007/BF02815377
Toda E, Koiso N, Takebayashi A, Ichikawa M, Kiba T, Osakabe K, Osakabe Y, Sakakibara H, Kato N, Okamoto T (2019) An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat Plants 5(4):4. https://doi.org/10.1038/s41477-019-0386-z
Tong L, Lei F-W, Wu Y-M, Shen X-L, Xia X-F, Zhang D-H, Mu X-Y, Zhang Z-X (2022) Multiple reproductive strategies of a spring ephemeral plant, Fritillaria maximowiczii, enable its adaptation to harsh environments. Plant Species Biol 37(1):38–51. https://doi.org/10.1111/1442-1984.12350
Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SBC, Nasmyth K (2000) Functional genomics identifies Monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell 103(7):7. https://doi.org/10.1016/S0092-8674(00)00217-8
Turnbull C, Lillemo M, Hvoslef-Eide TAK (2021) Global regulation of genetically modified crops amid the gene edited crop boom-a review. Front Plant Sci 12:244. https://doi.org/10.3389/fpls.2021.630396
Tzafrir I, McElver JA, Liu C, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW (2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128(1):38–51. https://doi.org/10.1104/pp.010911
Underwood CJ, Mercier R (2022) Engineering Apomixis: clonal seeds approaching the fields. Annu Rev Plant Biol 73(1):201–225. https://doi.org/10.1146/annurev-arplant-102720-013958
Underwood CJ, Vijverberg K, Rigola D, Okamoto S, Oplaat C, den Camp RHMO, Radoeva T, Schauer SE, Fierens J, Jansen K, Mansveld S, Busscher M, Xiong W, Datema E, Nijbroek K, Blom E-J, Bicknell R, Catanach A, Erasmuson S, van Dijk PJ (2022) A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat Genet 54(1):1. https://doi.org/10.1038/s41588-021-00984-y
van Dijk PJ, Bakx-Schotman JMT (2004) Formation of unreduced megaspores (Diplospory) in apomictic dandelions (Taraxacum officinale, s.l.) is controlled by a sex-specific dominant locus. Genetics 166(1):483–492. https://doi.org/10.1534/genetics.166.1.483
Van Dijk PJ, Op den Camp R, Schauer SE (2020) Genetic dissection of Apomixis in dandelions identifies a dominant parthenogenesis locus and highlights the complexity of autonomous endosperm formation. Genes 11(9):961. https://doi.org/10.3390/genes11090961
van Dijk PJ, Rigola D, Schauer SE (2016) Plant breeding: surprisingly, less sex is better. Curr Biol 26(3):R122–R124. https://doi.org/10.1016/j.cub.2015.12.010
van Dijk PJ, Tas ICQ, Falque M, Bakx-Schotman T (1999) Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83(6):6. https://doi.org/10.1046/j.1365-2540.1999.00620.x
Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, Laukens K, Pharazyn A, Ferreira PCG, Eloy N, Renne C, Meyer C, Faure J-D, De Jaeger G (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6(1):397. https://doi.org/10.1038/msb.2010.53
Vernet A, Meynard D, Lian Q, Mieulet D, Gibert O, Bissah M, Rivallan R, Autran D, Leblanc O, Meunier AC, Frouin J, Taillebois J, Shankle K, Khanday I, Mercier R, Sundaresan V, Guiderdoni E (2022) High-frequency synthetic apomixis in hybrid rice. Nat Commun 13(1):1. https://doi.org/10.1038/s41467-022-35679-3
Viana ML, Oliveira PE, Romero R, Caetano APS (2021) The best of both worlds: Apomixis and sexuality co-occur in species of Microlicia, Melastomataceae. Plant Species Biol 36(3):476–488. https://doi.org/10.1111/1442-1984.12330
Vijverberg K, Milanovic-Ivanovic S, Bakx-Schotman T, van Dijk PJ (2010) Genetic fine-mapping of DIPLOSPOROUS in Taraxacum (dandelion; Asteraceae) indicates a duplicated DIP-gene. BMC Plant Biol 10:154. https://doi.org/10.1186/1471-2229-10-154
Voigt-Zielinski M-L, Piwczyński M, Sharbel TF (2012) Differential effects of polyploidy and diploidy on fitness of apomictic Boechera. Sex Plant Reprod 25(2):97–109. https://doi.org/10.1007/s00497-012-0181-8
Vrielynck N, Chambon A, Vezon D, Pereira L, Chelysheva L, De Muyt A, Mézard C, Mayer C, Grelon M (2016) A DNA topoisomerase VI–like complex initiates meiotic recombination. Science 351(6276):939–943. https://doi.org/10.1126/science.aad5196
WFT, CAM, EWM, JLS, ACM, UELI, G, ARCO, BG, NINA C, BERNARDES DAJ, FREDERIQUE P (2016). Methods and compositions for the production of unreduced, non-recombined gametes and clonal offspring (WO 2016/179522 A1). https://lens.org/010-079-811-462-856
Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K (2011) The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol 21(15):1277–1281. https://doi.org/10.1016/j.cub.2011.07.001
Wang C, Liu Q, Shen Y, Hua Y, Wang J, Lin J, Wu M, Sun T, Cheng Z, Mercier R, Wang K (2019) Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol 37(3):283–286. https://doi.org/10.1038/s41587-018-0003-0
Wang N, Gent J, Dawe RK (2021) Haploid induction by a maize cenh3 null mutant. Sci Adv 7(4):eabe2299. https://doi.org/10.1126/sciadv.abe2299
Wang N, Song X, Ye J, Zhang S, Cao Z, Zhu C, Hu J, Zhou Y, Huang Y, Cao S, Liu Z, Wu X, Chai L, Guo W, Xu Q, Gaut BS, Koltunow AMG, Zhou Y, Deng X (2022) Structural variation and parallel evolution of apomixis in citrus during domestication and diversification. Natl Sci Rev 9(10):114. https://doi.org/10.1093/nsr/nwac114
Wang N, Xia X, Jiang T, Li L, Zhang P, Niu L, Cheng H, Wang K, Lin H (2022b) In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol J 20(1):22–24. https://doi.org/10.1111/pbi.13740
Wang X, Xu Y, Zhang S, Cao L, Huang Y, Cheng J, Wu G, Tian S, Chen C, Liu Y, Yu H, Yang X, Lan H, Wang N, Wang L, Xu J, Jiang X, Xie Z, Tan M, Xu Q (2017) Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet 49(5):5. https://doi.org/10.1038/ng.3839
Wang Y, Copenhaver GP (2018) Meiotic recombination: mixing it up in plants. Annu Rev Plant Biol 69(1):577–609. https://doi.org/10.1146/annurev-arplant-042817-040431
West M, Harada J (1993) Embryogenesis in higher-plants—an overview. Plant Cell 5(10):1361–1369
Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H (2006) The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol Biol Cell 17(3):1331–1343. https://doi.org/10.1091/mbc.e05-09-0902
Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim S-G, Kim S-T, Choe S, Kim J-S (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33(11):11. https://doi.org/10.1038/nbt.3389
Woodworth RH (1930) Cytological studies in the Betulaceae. III. Parthenogenesis and Polyembryony in Alnus rugosa. Botan Gazette 89(4):402–409. https://doi.org/10.1086/334072
Wu Z, Ji J, Tang D, Wang H, Shen Y, Shi W, Li Y, Tan X, Cheng Z, Luo Q (2015) OsSDS is essential for DSB formation in rice meiosis. Front Plant Sci 6:55. https://doi.org/10.3389/fpls.2015.00021
Xiao H, Luo H, Liu N, Turner C, Chen X, Ding H, Liang Y, Tan S, Tang J, Xiong D, Yang B (2021) High fruit setting rate without male participation: a case study of obligate apomixis in Rhomboda tokioi (Orchidaceae). Flora 283:151920. https://doi.org/10.1016/j.flora.2021.151920
Xue Z, Li Y, Zhang L, Shi W, Zhang C, Feng M, Zhang F, Tang D, Yu H, Gu M, Cheng Z (2016) OsMTOPVIB promotes meiotic DNA double-strand break formation in rice. Mol Plant 9(11):1535–1538. https://doi.org/10.1016/j.molp.2016.07.005
Yang C, Hamamura Y, Sofroni K, Böwer F, Stolze SC, Nakagami H, Schnittger A (2019) SWITCH 1/DYAD is a WINGS APART-LIKE antagonist that maintains sister chromatid cohesion in meiosis. Nat Commun 10(1):1. https://doi.org/10.1038/s41467-019-09759-w
Yao L, Zhang Y, Liu C, Liu Y, Wang Y, Liang D, Liu J, Sahoo G, Kelliher T (2018) OsMATL mutation induces haploid seed formation in indica rice. Nat Plants 4(8):530–533. https://doi.org/10.1038/s41477-018-0193-y
Yokobayashi S, Watanabe Y (2005) The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123(5):803–817. https://doi.org/10.1016/j.cell.2005.09.013
Yu H, Wang M, Tang D, Wang K, Chen F, Gong Z, Gu M, Cheng Z (2010) OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119(6):625–636. https://doi.org/10.1007/s00412-010-0284-7
Zhang C, Song Y, Cheng Z, Wang Y, Zhu J, Ma H, Xu L, Yang Z-N (2012) The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant J 72(2):271–281. https://doi.org/10.1111/j.1365-313X.2012.05075.x
Zhang Z, Conner J, Guo Y, Ozias-Akins P (2020) Haploidy in Tobacco induced byPsASGR-BBMLTransgenes via parthenogenesis. Genes 11(9):1072. https://doi.org/10.3390/genes11091072
Zhao X, Yuan K, Liu Y, Zhang N, Yang L, Zhang Y, Wang Y, Ji J, Fang Z, Han F, Lv H (2022) In vivo maternal haploid induction based on genome editing of DMP in Brassica oleracea. Plant Biotechnol J 20(12):2242–2244. https://doi.org/10.1111/pbi.13934
Zhong Y, Chen B, Li M, Wang D, Jiao Y, Qi X, Wang M, Liu Z, Chen C, Wang Y, Chen M, Li J, Xiao Z, Cheng D, Liu W, Boutilier K, Liu C, Chen S (2020) A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat Plants 6(5):466–472. https://doi.org/10.1038/s41477-020-0658-7
Zhong Y, Liu C, Qi X, Jiao Y, Wang D, Wang Y, Liu Z, Chen C, Chen B, Tian X, Li J, Chen M, Dong X, Xu X, Li L, Li W, Liu W, Jin W, Lai J, Chen S (2019) Mutation of ZmDMP enhances haploid induction in maize. Nat Plants 5(6):575–580. https://doi.org/10.1038/s41477-019-0443-7
Funding
Open Access funding enabled and organized by Projekt DEAL. This work is was supported by core funding from the Max Planck Society and the International Max Planck Research School on "Understanding Complex Plant Traits Using Computational and Evolutionary Approaches".
Author information
Authors and Affiliations
Contributions
AM, DKS and RM conceived the review. AM and DKS drafted the manuscript. AM, DKS and RM revised it.
Corresponding author
Ethics declarations
Conflict of interest
R. Mercier is listed as inventor in patents covering the use of MiMe to engineer apomixis.
Additional information
Communicated by David D Fang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahlandt, A., Singh, D.K. & Mercier, R. Engineering apomixis in crops. Theor Appl Genet 136, 131 (2023). https://doi.org/10.1007/s00122-023-04357-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04357-3