Abstract
Key message
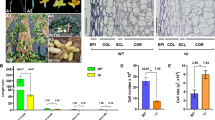
The novel spontaneous long hypocotyl and early flowering (lhef) mutation in cucumber is due to a 5551-bp LTR-retrotransposon insertion in CsPHYB gene encoding PHYTOCHROME B, which plays a major role in regulating photomorphogenic hypocotyl growth and flowering.
Abstract
Hypocotyl length and flowering time are important for establishing high-quality seedlings in modern cucumber production, but little is known for the underlying molecular mechanisms of these two traits. In this study, a spontaneous cucumber long hypocotyl and early flowering mutant was identified and characterized. Based on multiple lines of evidence, we show that cucumber phytochrome B (CsPHYB) is the candidate gene for this mutation, and a 5551-bp LTR-retrotransposon insertion in the first exon of CsPHYB was responsible for the mutant phenotypes. Uniqueness of the mutant allele at CsPHYB was verified in 114 natural cucumber lines. Ectopic expression of the CsPHYB in Arabidopsis phyB mutant rescued the long hypocotyl and early flowering phenotype of phyB-9 mutant. The wild-type CsPHYB protein was localized on the membrane and cytoplasm under white light condition, whereas in the nucleus under red light, it is consistent with its roles as a red-light photoreceptor in Arabidopsis. However, the mutant csphyb protein was localized on the membrane and cytoplasm under both white and red-light conditions. Expression dynamics of CsPHYB and several cell elongation-related genes were positively correlated with hypocotyl elongation; the transcription levels of key positive and negative regulators for flowering time were also consistent with the anthesis dates in the mutant and wild-type plants. Yeast two hybrid and bimolecular fluorescence complementation assays identified physical interactions between CsPHYB and phytochrome interacting factor 3/4 (CsPIF3/4). These findings will provide new insights into the roles of the CsPHYB in cucumber hypocotyl growth and flowering time.











Similar content being viewed by others
References
Blázquez MA, Weigel D (1999) Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol 120:1025–1032
Bo KL, Wang H, Pan YP, Behera TK, Pandey S, Wen CL, Wang YH, Simon PW, Li YH, Chen JF, Weng YQ (2016) SHORT HYPOCOTYL1 encodes a SMARCA3-like chromatin remodeling factor regulating elongation. Plant Physiol 172:1273–1292
Boeke JD, Corces VG (1989) Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43:403–434
Briggs WR, Olney MA (2001) Photoreceptors in plant photomorphogenesis to date: five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125:85–88
Cai YL, Bartholomew ES, Dong MM, Zhai XL, Yin S, Zhang YQ, Feng ZX, Wu LC, Liu W, Shan N, Zhang X, Ren HZ, Liu XW (2020) The HD-ZIP IV transcription factor GL2-Like regulates male flowering time and fertility in cucumber. J Exp Bot 71:5425–5437
Cavatorta J, Moriarty G, Glos M, Henning M, Kreitinger M, Mazourek M, Munger H, Jahn J (2012) Salt and pepper’: a disease-resistant cucumber Inbred. HortSci 47:427–428
Childs KL, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, Mullet JE (1997) The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol 113:611–619
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH (1992) COP1, an arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71:791–801
Devlin PF, Rood SB, Somers DE, Quail PH, Whitelam GC (1992) Photophysiology of the elongated internode (ein) mutant of brassica rapa: ein mutant lacks a detectable phytochrome B-like polypeptide. Plant Physiol 100:1442–1447
Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17:1941–1952
Fragoso V, Oh Y, Kim SG, Gase K, Baldwin IT (2017) Functional specialization of Nicotiana attenuata phytochromes in leaf development and flowering time. J Integr Plant Biol 59:205–224
Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61:11–24
Fukazawa J, Ohashi Y, Takahashi R, Nakai K, Takahashi Y (2021) DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 33:2258–2272
Galindo-González L, Mhiri C, Deyholos MK, Grandbastien MA (2017) LTR-retrotransposons in plants: engines of evolution. Gene 626:14–25
Galvāo VC, Fiorucci AS, Trevisan M, Franco-Zorilla JM, Goyal A, Schmid-Siegert E, Solano R, Fankhauser C (2019) PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nat Commun 10:4005
Ge XM, Hu X, Zhang J, Huang QM, Gao Y, Li ZQ, Li S, He JM (2020) UV RESISTANCE LOCUS8 mediates ultraviolet-B-induced stomatal closure in an ethylene-dependent manner. Plant Sci 301:110679
Hajdu A, Ádám É, Sheerin DJ, Dobos O, Bernula P, Hiltbrunner A, Kozma-Bognár L, Nagy F (2015) High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J 83:794–805
Heng YQ, Jiang Y, Zhao XH, Zhou H, Wang XC, Deng XW, Xu DQ (2019) BBX4, a phyB-interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc Natl Acad Sci USA 116:26049–26056
Hu LL, Liu P, Jin ZS, Sun J, Weng YQ, Chen P, Du SL, Wei AM, Li YH (2021) A mutation in CsHY2 encoding a phytochromobilin (PΦB) synthase leads to an elongated hypocotyl 1 (elh1) phenotype in cucumber (Cucumis sativus L.). Theor Appl Genet 134:2639–2652
Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21:2441–2450
Inoue SI, Kaiserli E, Zhao X, Waksman T, Takemiya A, Okumura M, Takahashi H, Seki M, Shinozaki K, Endo Y, Sawasaki T, Kinoshita T, Zhang X, Christie JM, Shimazaki KI (2020) CIPK23 regulates blue light-dependent stomatal opening in Arabidopsis thaliana. Plant J 104:679–692
Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD (2011) PFT1, the MED25 subunit of the plant mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J 69:601–612
Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH (2010) Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22:2370–2383
Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14:563–573
Kahle N, Sheerin DJ, Fischbach P, Koch LA, Schwenk P, Lambert D, Rodriguez R, Kerner K, Hoecker U, Zurbriggen MD, Hiltbrunner A (2020) COLD REGULATED 27 and 28 are targets of CONSTITUTIVELY PHOTOMORPHOGENIC 1 and negatively affect phytochrome B signalling. Plant J 104:1038–1053
Kippes N, Vangessel C, Hamilton J, Akpinar A, Budak H, Dubcovsky J, Pearce S (2020) Effects of phyB and phyC loss-of-function mutations on the wheat transcriptome under short and long day photoperiods. BMC Plant Biol 20:297
Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100:147–160
Kumar A, Singh A, Panigrahy M, Sahoo PK, Panigrahi-Kishore CS (2018) Carbon nanoparticles influence photomorphogenesis and flowering time in Arabidopsis thaliana. Plant Cell Rep 37:901–912
Lazaro A, Mouriz A, Piñeiro M, Jarillo JA (2015) Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27:2437–2454
Lee IJ, Foster KR, Morgan PW (1998a) Photoperiod control of gibberellin levels and flowering in sorghum. Plant Physiol 116:1003–1011
Lee IJ, Foster KR, Morgan PW (1998b) Effect of gibberellin biosynthesis inhibitors on native gibberellin content, growth and floral initiation in Sorghum bicolor. J Plant Growth Regul 17:185–195
Lee YS, Jeong DH, Lee DY, Yi J, Ryu CH, Kim SL, Jeong HJ, Choi SC, Jin P, Yang J, Cho LH, Choi H, An G (2010) OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J 63:18–30
Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16:19–28
Li XY (2011) Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protoc 1:1–3
Li YH, Wen CL, Weng YQ (2013) Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor Appl Genet 126:2187–2196
Liu B, Weng JY, Guan DL, Zhang Y, Niu QL, López-Juez E, Lai YS, Garcia-Mas J, Huang DF (2021a) A domestication-associated gene, CsLH, encodes a phytochrome B protein that regulates hypocotyl elongation in cucumber. Mol Hortic 1:3
Liu SR, Yang LW, Li JL, Tang WJ, Li JG, Lin RC (2021b) FHY3 interacts with phytochrome B and regulates seed dormancy and germination. Plant Physiol 187:289–302
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Lu HF, Lin T, Klein J, Wang SH, Qi JJ, Zhou Q, Sun JJ, Zhang ZH, Weng YQ, Huang SW (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127:1491–1499
Lu X, Zhou C, Xu P, Luo Q, Lian H, Yang H (2015) Red light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and phoeomorphogenic development in Arabidopsis. Mol Plant 8:467–478
Mao ZL, He SB, Xu F, Wei XX, Jiang L, Wang WX, Li T, Xu PB, Du SS, Li L, Lian HL, Guo TT, Yang HQ (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225:848–865
Miao TT, Li DZ, Huang ZY, Huang YW, Li SS, Wang Y (2021) Gibberellin regulates UV-B-induced hypocotyl growth inhibition in Arabidopsis thaliana. Plant Signal Behav 16:1966587
Miao LX, Zhao JC, Yang GQ, Xu P, Cao XL, Du SS, Xu F, Jiang L, Zhang SL, Wei XX, Liu Y, Chen HR, Mao ZL, Guo TT, Kou S, Wang WX, Yang HQ (2022) Arabidopsis cryptochrome 1 undergoes COP1 and LRBs-dependent degradation in response to high blue light. New Phytol 234:1347–1362
Ming CH, Jiang FL, Hu HM, Zhou XC, Zhan FH, Wu Z (2011) Effects of different leggy extent seedling on cucumber growth, yield and quality. China Veg 4:29–34 (In Chinese)
Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126:2073–2082
Nagatani A (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13:565–570
Nakano T (2019) Hypocotyl elongation: a molecular mechanism for the first event in plant growth that influences its physiology. Plant Cell Physiol 60:933–934
Nemhauser J, Chory J (2002) Photomorphogenesis the Arabidopsis. Book 1:e0054
Oda A, Fujiwara S, Kamada H, Coupland G, Mizoguchi T (2004) Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett 557:259–264
Oh J, Park E, Song K, Bae G, Choi G (2020) Phytochrome interacting factor 8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis. Plant Cell 32:186–205
Pan Y, Qu SP, Bo KL, Gao ML, Haider KR, Weng Y (2017) QTL mapping of domestication and diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor Appl Genet 130:1531–1548
Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J (2016) RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol 16:141
Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3:85–93
Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5:147–157
Ren Y, Zhang Z, Liu J, Staub JE, Han Y, Cheng Z, Li X, Lu J, Miao H, Kang H, Xie B, Gu X, Wang X, Du C, Jin W, Huang S (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 4:e5795
Robbins MD, Staub JE (2009) Comparative analysis of marker-assisted and phenotypic selection for yield components in cucumber. Theor Appl Genet 119:621–634
Rusaczonek A, Czarnocka W, Willems P, Sujkowska-Rybkowska M, Breusegem FN, Karpiński S (2021) Phototropin 1 and 2 influence photosynthesis, UV-C induced photooxidative stress responses, and cell death. Cells 10:200
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sheerin DJ, Menon C, Oven-Krockhaus SZ, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with memebers of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27:189–201
Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E (2008) Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20:1586–1602
Sheng YY, Pan YP, Li YH, Yang LM, Weng YQ (2019) Quantitative trait loci for fruit size and flowering time-related traits under domestication and diversifying selection in cucumber (Cucumis sativus). Plant Breeding 139:176–191
Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66:441–464
Su L, Hou P, Song MF, Zheng X, Guo L, Xiao Y, Yan L, Li WC, Yang JP (2015) Synergistic and antagonistic action of phytochrome (Phy) A and PhyB during seedling de-etiolation in Arabidopsis thaliana. Int J Mol Sci 16:12199–12212
Sun WJ, Han HY, Deng L, Sun CL, Xu TR, Li LH, Ren PR, Zhao JH, Zhai QZ, Li CY (2020) Mediator subunit MED25 physically interacts with PHYTOCHROME INTERACTING FACTOR4 to regulate shade-induced hypocotyl elongation in tomato. Plant Physiol 184:1549–1562
Suzuki A, Suriyagoda L, Shigeyama T, Tominaga A, Sasaki M, Hiratsuka Y, Yoshinaga A, Arima S, Agarie S, Sakai T, Inada S, Jikumaru Y, Kamiya Y, Uchiumi T, Abe M, Hashiguchi M, Akashi R, Sato S, Kaneko T, Tabata S, Hirsh AM (2011) Lotus japonicus nodulation is photomorphogenetically controlled by sensing the red/far red (R/FR) ration through jasmonic acid (JA) signaling. Proc Natl Acad Sci USA 108:16837–16842
Takano M, Inagaki N, Xie XZ, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of de-etiolation and flowering in rice. Plant Cell 17:3311–3325
Tuinen AV, Kerckhoffs LHJ, Nagatani A, Kendrick RE, Koornneef M (1995) A temporarily red light-insensitive mutant of tomato lacks a light-stable, B-like phytochrome. Plant Physiol 108:939–947
Wagner D, Koloszvari M, Quail PH (1996) Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8:859–871
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wang YH, Bo KL, Gu XF, Pan JS, Li YH, Chen JF, Wen CL, Ren ZH, Ren HZ, Chen XH, Grumet G, Weng Y (2019) Molecularly tagged genes and quantitative trait loci in cucumber with recommendations for QTL nomenclature. Hortic Res 7:3
Wei XX, Wang WT, Xu P, Wang WX, Guo TT, Kou S, Liu MQ, Niu YK, Yang HQ, Mao ZL (2021) Phytochrome B interacts with SWC6 and ARP6 to regulate H2A.Z deposition and photomorphogenesis in Arabidopsis. J Integr Plant Biol 63:1133–1146
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, Sanmiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wollenberg AC, Strasser B, Cerdán PD, Amasino RM (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148:1681–1694
Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006
Xu PB, Lian HL, Xu F, Zhang T, Wang S, Wang WX, Du SS, Huang JR, Yang HQ (2019) Phytochrome B and AGB1 coordinately regulate photomorphogenesis by antagonistically modulating PIF3 stability in Arabidopsis. Mol Plant 12:229–247
Xu P, Chen HR, Li T, Xu F, Mao ZL, Cao XL, Miao LX, Du SS, Hua J, Zhao JC, Guo TT, Kou S, Wang WX, Yang HQ (2021) Blue light-dependent interactions of CRY1 with GID1 and DELLA proteins regulate gibberellin signaling and photomorphogenesis in Arabidopsis. Plant Cell 33:2375–2394
Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK, Datta S (2020) Light signaling and UV-B-mediated plant growth regulation. J Integr Plant Biol 62:1270–1292
Yan Y, Li C, Dong XJ, Li H, Zhang D, Zhou YY, Jiang BC, Peng J, Qin XY, Cheng JK, Wang XJ, Song PY, Qi LJ, Zheng Y, Li BS, Terzaghi W, Yang SH, Guo Y, Li JG (2020) MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32:2196–2215
Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T (2012) The phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 AND VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 24:3248–3263
Yoshida Y, Sarmiento-Mañús R, Yamori W, Ponce MR, Micol JL, Tsukaya H (2018) The Arabidopsis phyB-9 mutant has a second-site mutation in the VENOSA4 gene that alters chloroplast size, photosynthetic traits, and leaf growth. Plant Physiol 178:3–6
Zhong M, Zeng BJ, Tang DY, Yang JX, Qu LN, Yan JD, Wang XC, Li X, Liu XM, Zhao XY (2021) The blue light receptor CRY1 interacts with GID1 and DELLA proteins to repress GA signaling during photomorphogenesis in Arabidopsis. Mol Plant 14:1328–1342
Zou Y, Li R, Baldwin IT (2020) ZEITLUPE is required for shade avoidance in the wild tobacco Nicotiana attenuata. J Integr Plant Biol 62:1341–1351
Acknowledgements
Work in PC’s and HY’s lab was supported by the National Natural Science Foundation of China under Project #31860557. YL’s lab was supported by the National Natural Science Foundation of China under Project #31772300. YW’s lab was supported by the Agriculture and Food Research Initiative competitive Grant No. 2017-67013-26195 of the USDA National Institute of Food and Agriculture.
Author information
Authors and Affiliations
Contributions
LH performed the research and prepared a draft of the manuscript. MZ, JS, ZL and YW participated in the research. HY participated in data analysis and provided technical help. YL and PC designed the experiments, supervised this study and revised the manuscript with inputs from YW. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Data availability
All data generated or analyzed that support the findings of this study are included in the main text article and its supplementary files.
Additional information
Communicated by Richard G.F. Visser.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, L., Zhang, M., Shang, J. et al. A 5.5-kb LTR-retrotransposon insertion inside phytochrome B gene (CsPHYB) results in long hypocotyl and early flowering in cucumber (Cucumis sativus L.). Theor Appl Genet 136, 68 (2023). https://doi.org/10.1007/s00122-023-04271-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04271-8