Abstract
Key message
High-density haplotype analysis revealed significant haplotype sharing between ex-PVPs registered from 1976 to 1992 and key maize founders, and uncovered similarities and differences in haplotype sharing patterns by company and heterotic group.
Abstract
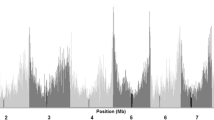
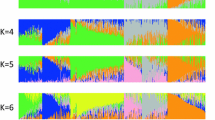
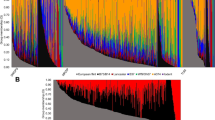
Proprietary inbreds developed by the private seed industry have been the major source for driving genetic gain in successful North American maize hybrids for decades. Much of the history of industry germplasm can be traced back to key founder lines, some of which were pivotal in the development of prominent heterotic groups. Previous studies have summarized pedigree-based relationships, genetic diversity and population structure among commercial inbreds with expired Plant Variety Protection (ex-PVP). However, less is known about the extent of haplotype sharing between historical founders and ex-PVPs. A better understanding of the relationships between founders and ex-PVPs provides insight into the haplotype and heterotic group structure among industry germplasm. We performed high-density haplotype analysis with 11.3 million SNPs on 212 maize inbreds, which included 157 ex-PVPs registered 1976–1992 and 55 public lines relevant to PVPs. Among these lines were 12 key founders identified in literature review: 207, A632, B14, B37, B73, LH123HT, LH82, Mo17, Oh43, OH7, PHG39 and Wf9. Our results revealed that, on average, 81.6% of an ex-PVP’s genome is shared with at least 1 of these 12 founder lines and more than half when limited to B73, Mo17 and 207. Quantifiable similarities and contrasts among heterotic groups and major US seed industry companies were also observed. The results from this study provide high-resolution haplotype data on ex-PVP germplasm, confirm founder relationship trends observed in previous studies, uncover region-specific haplotype structure differences and demonstrate how haplotype sharing analysis can be used as a tool to explore germplasm diversity.







Similar content being viewed by others
Data availability
File S2 is available for download at https://figshare.com/s/b48a85564d5c29f778da.
References
Beckett TJ, Morales AJ, Koehler KL, Rocheford TR (2017) Genetic relatedness of previously plant-variety-protected commercial maize inbreds. PLoS ONE. https://doi.org/10.1371/journal.pone.0189277
Bernardo R (1993) Estimation of coefficient of coancestry using molecular markers in maize. Theor Appl Genet 85(8):1055–1062. https://doi.org/10.1007/BF00215047
Bernardo R, Romero-Severson J, Ziegle J, Hauser J, Joe L, Hookstra G, Doerge RW (2000) Parental contribution and coefficient of coancestry among maize inbreds: pedigree, RFLP, and SSR data. Theor Appl Genet 100(3–4):552–556. https://doi.org/10.1007/s001220050072
Bogenschutz TG, Russell WA (1986) An evaluation for genetic variation within maize inbred lines maintained by sib-mating and self-pollination. Euphytica 35(2):403–412. https://doi.org/10.1007/BF00021848
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17(2):343–360. https://doi.org/10.1105/tpc.104.025627
Bukowski R, Guo X, Lu Y et al (2015) Construction of the third generation Zea mays haplotype map. bioRxiv. https://doi.org/10.1101/026963
Bukowski R, Guo X, Lu Y et al (2018) Construction of the third-generation Zea mays haplotype map. GigaScience. https://doi.org/10.1093/gigascience/gix134
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP ++. PLoS Comput Biol 6(12):e1001025. https://doi.org/10.1371/journal.pcbi.1001025
Dell’Acqua M, Gatti DM, Pea G et al (2015) Genetic properties of the MAGIC maize population: a new platform for high definition QTL mapping in Zea mays. Genome Biol 16:167. https://doi.org/10.1186/s13059-015-0716-z
East EM (1908) Inbreeding in corn. Conn Agric Exp Stn Rep 1907:419–428
Fang Z, Gonzales AM, Clegg MT et al (2014) Two genomic regions contribute disproportionately to geographic differentiation in wild barley. G3 Genes Genomes Genet 4(7):1193–1203. https://doi.org/10.1534/g3.114.010561
Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner E, Hansen M, Joets J, Paslier ML, McMullen M, Montalent P, Rose M, Schön CC, Sun Q, Walter H, Martin O, Falque M (2011) A large Maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6(12):e28334
Gattepaille LM, Jakobsson M (2012) Combining markers into haplotypes can improve population structure inference. Genetics 190(1):159–174. https://doi.org/10.1534/genetics.111.131136
Gethi JG, Labate JA, Lamkey KR, Smith ME, Kresovich S (2002) SSR variation in Important U.S. maize inbred lines. Crop Sci 42(3):951–957. https://doi.org/10.2135/cropsci2002.9510
Haasl RJ, Payseur BA (2011) Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity 106(1):158–171. https://doi.org/10.1038/hdy.2010.21
Haun WJ, Hyten DL, Xu WW et al (2011) The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol 155(2):645–655. https://doi.org/10.1104/pp.110.166736
Hirsch C, Hirsch CD, Brohammer AB et al (2016) Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell. https://doi.org/10.1105/tpc.16.00353
Hufford MB, Xu X, van Heerwaarden J et al (2012) Comparative population genomics of maize domestication and improvement. Nat Genet 44(7):808–811. https://doi.org/10.1038/ng.2309
Hufford MB, Lubinksy P, Pyhäjärvi T, Devengenzo MT, Ellstrand NC, Ross-Ibarra J (2013) The genomic signature of crop-wild introgression in maize. PLoS Genet 9(5):e1003477. https://doi.org/10.1371/journal.pgen.1003477
Inghelandt DV, Melchinger AE, Lebreton C, Stich B (2010) Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor Appl Genet 120(7):1289–1299. https://doi.org/10.1007/s00122-009-1256-2
Jiao Y, Zhao H, Ren L et al (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44(7):812–815. https://doi.org/10.1038/ng.2312
Johnson LC, Bradbury P, Casstevens T et al (2018) A practical haplotype graph for determining genomic sequence. In: Poster presented at the plant & animal genome conference XXVI, San Diego, CA
Kahler AL, Kahler JL, Thompson SA et al (2010) North American study on essential derivation in maize: II. Selection and evaluation of a panel of simple sequence repeat loci. Crop Sci 50(2):486. https://doi.org/10.2135/cropsci2009.03.0121
Kaufman L, Rousseeuw PJ (2008) Partitioning around medoids (program PAM). In: Finding groups in data: an introduction to cluster analysis. Wiley, Hoboken. Retrieved from http://onlinelibrary.wiley.com/doi/10.1002/9780470316801.ch2/summary
Kebede AZ, Woldemariam T, Reid LM, Harris LJ (2016) Quantitative trait loci mapping for Gibberella ear rot resistance and associated agronomic traits using genotyping-by-sequencing in maize. Theor Appl Genet 129(1):17–29. https://doi.org/10.1007/s00122-015-2600-3
Kremling KAG, Chen S-Y, Su M-H et al (2018) Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature. https://doi.org/10.1038/nature25966
Lai J, Li R, Xu X et al (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42(11):1027–1030. https://doi.org/10.1038/ng.684
Lawson DJ, Hellenthal G, Myers S, Falush D (2012) Inference of population structure using dense haplotype data. PLoS Genet 8(1):e1002453. https://doi.org/10.1371/journal.pgen.1002453
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Liang Z, Schnable JC (2016) RNA-seq based analysis of population structure within the maize inbred B73. PLoS ONE 11(6):e0157942. https://doi.org/10.1371/journal.pone.0157942
Lorenz A, Hoegemeyer T (2013) The phylogenetic relationships of U.S. maize germplasm. Nat Genet 45(8):844–845. https://doi.org/10.1038/ng.2697
Lu Y, Zhang S, Shah T et al (2010) Joint linkage–linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc Natl Acad Sci 107(45):19585–19590. https://doi.org/10.1073/pnas.1006105107
Lu F, Romay MC, Glaubitz JC et al (2015) High-resolution genetic mapping of maize pan-genome sequence anchors. Nat Commun. https://doi.org/10.1038/ncomms7914
Lübberstedt T, Melchinger AE, Dußle C, Vuylsteke M, Kuiper M (2000) Relationships among early European maize inbreds: IV. Genetic diversity revealed with AFLP markers and comparison with RFLP, RAPD, and pedigree data. Crop Sci 40(3):783–791. https://doi.org/10.2135/cropsci2000.403783x
Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K (2018) cluster: cluster analysis basics and extensions
Mangelsdorf PC (1975) Donald Forsha Jones (1890–1963) In: Biographical memoirs, vol 46, pp 135–156. National Academy of Science. Retrieved from https://doi.org/10.17226/569
Messmer MM, Melchinger AE, Herrmann RG, Boppenmaier J (1993) Relationships among early European maize inbreds: II. Comparison of pedigree and RFLP data. Crop Sci 33(5):944–950. https://doi.org/10.2135/cropsci1993.0011183X003300050014x
Mikel MA (2008) Genetic diversity and improvement of contemporary proprietary North American dent corn. Crop Sci 48(5):1686. https://doi.org/10.2135/cropsci2008.01.0039
Mikel MA (2011) Genetic composition of contemporary U.S. commercial dent corn germplasm. Crop Sci 51(2):592–599. https://doi.org/10.2135/cropsci2010.06.0332
Mikel MA, Dudley JW (2006) Evolution of North American dent corn from public to proprietary germplasm. Crop Sci 46(3):1193. https://doi.org/10.2135/cropsci2005.10-0371
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, Columbia
Nelson PT, Coles ND, Holland JB, Bubeck DM, Smith S, Goodman MM (2008) Molecular characterization of maize inbreds with expired U.S. plant variety protection. Crop Sci 48(5):1673–1685. https://doi.org/10.2135/cropsci2008.02.0092
Nelson PT, Krakowsky MD, Coles ND, Holland JB, Bubeck DM, Smith JSC, Goodman MM (2016) Genetic characterization of the North Carolina State University maize lines. Crop Sci 56(1):259. https://doi.org/10.2135/cropsci2015.09.0532
Paten B, Novak AM, Eizenga JM, Garrison E (2017) Genome graphs and the evolution of genome inference. Genome Res 1:2–3. https://doi.org/10.1101/gr.214155.116
Poets AM, Mohammadi M, Seth K et al (2016) The effects of both recent and long-term selection and genetic drift are readily evident in North American barley breeding populations. G3 Genes Genomes Genet 6(3):609–622. https://doi.org/10.1534/g3.115.024349
Raj A, Stephens M, Pritchard JK (2014) fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197(2):573–589. https://doi.org/10.1534/genetics.114.164350
Ralph P, Coop G (2013) The geography of recent genetic ancestry across Europe. PLoS Biol 11(5):e1001555. https://doi.org/10.1371/journal.pbio.1001555
Ramu P, Esuma W, Kawuki R et al (2017) Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat Genet 49(6):959–963. https://doi.org/10.1038/ng.3845
Romay MC, Millard MJ, Glaubitz JC et al (2013) Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol 14:R55. https://doi.org/10.1186/gb-2013-14-6-r55
Romero-Severson J, Smith JSC, Ziegle J et al (2001) Pedigree analysis and haplotype sharing within diverse groups of Zea mays L. inbreds. Theor Appl Genet 103(4):567–574. https://doi.org/10.1007/PL00002911
Ros-Freixedes R, Gonen S, Gorjanc G, Hickey JM (2017) A method for allocating low-coverage sequencing resources by targeting haplotypes rather than individuals. Genet Sel Evol 49:78. https://doi.org/10.1186/s12711-017-0353-y
Shull GH (1908) The composition of a field of maize. J Hered 4(1):296–301. https://doi.org/10.1093/jhered/os-4.1.296
Shull GH (1914) Duplicate genes for capsule form in bursa bursa-pastoris. Z Indukt Abstammungs u Verbungsl 12:97–149
Smith S (2007) Pedigree background changes in U.S. hybrid maize between 1980 and 2004. Crop Sci 47(5):1914–1926. https://doi.org/10.2135/cropsci2006.12.0763
Smith JSC, Duvick DN, Smith OS, Grunst A, Wall SJ (1999) Effect of hybrid breeding on genetic diversity in maize. In: Coors JG, Pandey S (eds) The genetics and exploitation of heterosis in crops, vol acsesspublicati, pp 119–126. Crop Science Society of America, Madison. Retrieved from https://dl-sciencesocieties-org.proxy.lib.iastate.edu/publications/books/articles/acsesspublicati/thegeneticsande/119
Springer NM, Ying K, Fu Y et al (2009) Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet 5(11):e1000734. https://doi.org/10.1371/journal.pgen.1000734
Swarts K, Li H, Navarro R et al (2014) Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome. https://doi.org/10.3835/plantgenome2014.05.0023
Technow F, Schrag TA, Schipprack W, Melchinger AE (2014) Identification of key ancestors of modern germplasm in a breeding program of maize. Theor Appl Genet 127(12):2545–2553. https://doi.org/10.1007/s00122-014-2396-6
Tracy WF, Chandler MA (2006) The historical and biological basis of the concept of heterotic patterns in corn belt dent maize. In: Lamkey KR, Lee M (eds) Plant breeding: the Arnel R. Hallauer international symposium, pp 219–233. Blackwell Publishing. Retrieved from http://onlinelibrary.wiley.com/doi/10.1002/9780470752708.ch16/summary
Troyer AF (1999) Background of U.S. hybrid corn. Crop Sci 39(3):601–626. https://doi.org/10.2135/cropsci1999.0011183X003900020001x
van Heerwaarden J, Hufford MB, Ross-Ibarra J (2012) Historical genomics of North American maize. Proc Natl Acad Sci USA 109(31):12420–12425. https://doi.org/10.1073/pnas.1209275109
Wang L, Beissinger TM, Lorant A, Ross-Ibarra C, Ross-Ibarra J, Hufford MB (2017) The interplay of demography and selection during maize domestication and expansion. Genome Biol. https://doi.org/10.1186/s13059-017-1346-4
Wu X, Li Y, Fu J et al (2016) Exploring identity-by-descent segments and putative functions using different foundation parents in maize. PLoS ONE 11(12):e0168374. https://doi.org/10.1371/journal.pone.0168374
Yang J, Mezmouk S, Baumgarten A et al (2017) Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007019
Zhou X, Carter TE, Cui Z, Miyazaki S, Burton JW (2000) Genetic base of Japanese soybean cultivars released during 1950 to 1988. Crop Sci 40(6):1794–1802. https://doi.org/10.2135/cropsci2000.4061794x
Acknowledgements
The author SMC wishes to thank Justin Gerke and Dean Podlich for helpful discussions in preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
SMC designed the study, designed and ran the analyses, interpreted the results and wrote the manuscript. MBH, CMA and TL assisted with the study design, interpretation of results and provided critical feedback during manuscript preparation. TL supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The author SMC is employed by Corteva Agriscience™, Agriculture Division of DowDuPont™. The funder provided support in the form of salary and graduate program support for author SMC, but did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The specific roles of this author are articulated in the author contribution statement. The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that ethical standards are met, and all the experiments comply with the current laws of the country in which they were performed.
Additional information
Communicated by Alain Charcosset.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Coffman, S.M., Hufford, M.B., Andorf, C.M. et al. Haplotype structure in commercial maize breeding programs in relation to key founder lines. Theor Appl Genet 133, 547–561 (2020). https://doi.org/10.1007/s00122-019-03486-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03486-y