Abstract
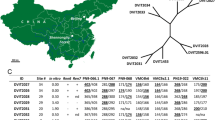
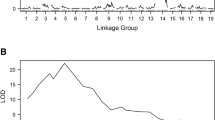
Pierce’s disease (PD) limits the cultivation of Vitis vinifera grape cultivars in California, across the southern United States and into South America. Resistance has been well characterized in V. arizonica, and one resistance locus has been identified (PdR1). However, resistance is poorly characterized in most other grape species. We tested a wide range of Vitis species from the southwestern United States for resistance to PD and used nuclear and chloroplast markers to phenotypically and genetically select a diverse set of resistant accessions. Chloroplast SSR markers identified 11 maternal lineage lines within the set of 17 (14 new and three previously identified) PD resistant accessions. A total of 19 breeding populations (F1 and pseudo-BC1) were developed with the 14 PD resistant accessions, and a total of 705 seedlings were analyzed for PD resistance. Using a limited mapping approach, 12 SSR markers, linked to the PdR1 locus, were used to genotype the breeding populations and phenotypic data were analyzed. Nine accessions had a major resistance quantitative trait locus (QTL) within the genomic region containing PdR1. The phenotypic data for these three resistant accessions, ANU67, b41-13, and T03-16, did not associate with PdR1 linked markers, indicating that their resistance is located in other regions of the genome. These three accessions were identified as candidates for use in the development of framework maps with larger populations capable of detecting additional and unique loci for PD resistance breeding and the stacking of PD resistance genes.



Similar content being viewed by others
References
Almeida RPP, Purcell AH (2003) Biological traits of Xylella fastidiosa strains for grapes and almonds. Appl Environ Microbiol 69:7447–7452
Arroyo-Garcia R, Lefort F, de Andrés MT, Ibañez J, Borrego J, Jouve N, Cabello F, Martinez-Zapater JM (2002) Chloroplast microsatellite polymorphisms in Vitis species. Genome 45:1142–1149
Beretta MJG, Barthe GA, Ceccardi TL, Lee RF, Derrich KS (1997) A survey for strains of Xylella fastidiosa in citrus affected by citrus variegated chlorosis and citrus blight in Brazil. Plant Dis 81:1196–1198
Bhattacharyya A, Stilwagen S, Ivanova N, D’Souza M, Bernal A, Lykidis A, Kapatral V, Anderson I, Larsen N, Los T, Reznik G, Selkov E Jr, Walunas TL, Feil H, Feil WS, Purcell A, Lassez JL, Hawkins TL, Haselkorn R, Overbeek R, Predki PF, Kyrpides NC (2002) Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc Natl Acad Sci USA 99:12403–12408
Bowers J, Boursiquot JM, This P, Chu K, Johansson H, Meredith C (1999) Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science 285:1562–1565
Chatterjee S, Almeida RPP, Lindow S (2008) Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol 46:243–271
Clarke EE, Ren Z, Lu J (2003) Evaluation of grape germplasm for resistance to Pierce’s disease and glassy-winged sharpshooter. Proc Fla State Hortic Soc 116:32–35
Crall JM, Stover LH (1957) The significance of Pierce’s disease in the decline of bunch grapes in Florida. Phytopathology 47:518
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341:746–751
Elbeaino T, Valentini F, Abou Kubaa R, Moubarak P, Yaseen T, Digiaro M (2014) Multilocus sequence typing of Xylella fastidiosa isolated from olive affected by “olive quick decline syndrome” in Italy. Phytopathol Mediterr 53:533–542
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol 13:39
Goheen AC, Raju BC, Lowe SK, Nyland G (1979) Pierce’s disease of grapevines in Central America. Plant Dis Rep 63:788–793
Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011
Hernandez-Martinez R, de la Cerda KA, Costa HS, Cooksey DA, Wong FP (2007) Phylogenetic relationships of Xylella fastidiosa strains isolated from landscape ornamentals in Southern California. Phytopathology 97:857–864
Hewitt WB (1958) The probable home of Pierce’s disease virus. Plant Dis Report 42:211–215
Hopkins DL (1980) Use of pin-prick inoculation technique demonstrates variability in virulence of Pierce’s disease bacterium. In: Proceedings of the 7th international conference on viruses grapevines (ICVG), Niagara Falls, pp 177–180
Hopkins DL, Purcell AH (2002) Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis 86:1056–1066
Hopkins DL, Mollenhauer HH, Mortensen JA (1974) Tolerance to Pierce’s disease and the associated rickettsia-like bacterium in muscadine grape. J Am Soc Hortic Sci 99:436–439
Ibanez J, Velez MD, de Andrés MT, Borrego J (2009) Molecular markers for establishing distinctness in vegetatively propagated crops: a case study in grapevine. Theor Appl Genet 119:1213–1222
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jimenez A (1985) Immunological evidence of Pierce’s disease of grapevine in Venezuela. Turrialba 35:243–247
Krivanek AF, Walker MA (2005) Vitis resistance to Pierce’s disease is characterized by differential Xylella fastidiosa populations in stems and leaves. Phytopathology 95:44–52
Krivanek AF, Riaz S, Walker MA (2006) Identification and molecular mapping of PdR1, a primary resistance gene to Pierce’s disease in Vitis. Theor Appl Genet 112:1125–1131
Kung SH, Almeida RPP (2011) Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 77:5278–5284
Lacombe T, Boursiquot J-M, Laucou V, Di Vecchi-Staraz M, Pe’ros J-P, This P (2013) Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor Appl Genet 126:401–414
Laucou V, Lacombe T, Dechesne F, Siret R, Bruno JP, Dessup M, Dessup T, Ortigosa P, Parra P, Roux C, Santoni S, Varès D, Péros JP, Boursiquot JM, This P (2011) High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122:1233–1245
Li WB, Pria WD, Teixeira C, Miranda VS, Ayres AJ, Franco CF, Costa MG (2001) Coffee leaf scorch caused by a strain of Xylella fastidiosa from citrus. Plant Dis 85:501–505
Loconsole G, Potere O, Boscia D, Altamura G, Djelouah K, Elbeaino T et al (2014) Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J Plant Pathol 96:7–14
Loomis NH (1958) Performance of Vitis species in the south as an indication of their relative resistance to Pierce’s disease. Plant Dis Rep 42:833–836
Mortensen JA (1968) The inheritance of resistance to Pierce’s disease in Vitis. J Am Soc Hortic Sci 92:331–337
Mortensen JA (1988) Blanc Du Bois grape. Hortic Sci 23:418–419
Mortensen JA, Stover LH (1982) Tampa grape rootstock. HortScience 17:273–274
Mortensen JA, Stover LH, Balerdi CF (1977) Sources of resistance to Pierce’s disease in Vitis. J Am Soc Hortic Sci 102:695–697
Mysore KS, Ryu CM (2004) Nonhost resistance: how much do we know? Trend Plant Sci 9:97–104
Nome SF, Docampo D, Goheen AC, Raju BC, Nyland G (1981) An enzyme-linked immunosorbent assay (ELISA) for detection of Pierce’s disease bacteria in plant tissues. Phytopathology 71:107
Nunney L, Yuan X, Bromley R, Hartung J, Montero-Astua M, Moreira L, Ortiz B, Stouthamer R (2010) Population genomic analysis of bacterial plant pathogen: novel insight into the origin of Pierce’s disease of grapevine in the U.S. PLoS ONE 11:1–9
Nunney L, Schuenzel EL, Scally M, Bromley RE, Stouthamer R (2014) Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl Environ Microbiol 80:3025–3033
Overcash J, Hegwood C, Stojanovic B (1981) ‘Midsouth’ and ‘Missblue’—two new bunch grape cultivars. In: Mississippi Agricultural and Forestry Experiment Station research report, vol 6, issue 18
Péros JP, Berger G, Portemont A, Boursiquot JM, Lacombe T (2011) Genetic variation and biogeography of the disjunct Vitis subgenus (Vitaceae). J Biogeogr 38(3):471–486
Perrier X, Jacquemoud-'Collet JP (2006) DARwin software. http://darwin.cirad.fr/. Accessed 25 Apr 2018
Perry RL, Mollenhauer HH, Bowen HH (1974) Electron photo-microscopy verification of Pierce’s disease on grape plants from Texas. Plant Dis Rep 58:780–782
Pierce N (1892) The California vine disease. US Dep Agric Div Veg Pathol Bull 2:222p
Provan J, Soranzo N, Wilson NJ, Goldstein DB, Powell W (1999) A low mutation rate for chloroplasts microsatellites. Genetics 153:943–947
Purcell AH, Saunders SR (1999) Fate of Pierce’s disease strains of Xylella fastidiosa in common riparian plants in California. Plant Dis 83:825–830
Randall JJ, Goldberg NP, Kemp JD, Radionenko M, French JM, Olsen MW et al (2009) Genetic analysis of a novel Xylella fastidiosa subspecies found in the southwestern United States. Appl Environ Microbiol 75:5631–5638
Riaz S, Krivanek AF, Xu K, Walker MA (2006) Refined mapping of the Pierce’s disease resistance locus, PdR1, and sex on an extended genetic map of Vitis rupestris × Vitis arizonica. Theor Appl Genet 113:1317–1329
Riaz S, Vezzulli S, Harbertson ES, Walker MA (2007) Use of molecular markers to correct grape breeding errors and determine the identity of novel sources of resistance to Xiphinema index and Pierce’s disease. Am J Enol Vitic 58:494–498
Riaz S, Tenscher AC, Rubin J, Graziani R, Pao SS, Walker MA (2008) Fine-scale genetic mapping of two Pierce’s disease resistance loci and a major segregation distortion region on chromosome 14 of grape. Theor Appl Genet 117:671–681
Riaz S, Tenscher A, Graziani R, Krivanek A, Ramming D, Walker A (2009) Using marker-assisted selection to breed Pierce’s disease-resistant grapes. Am J Enol Vitic 60:199–207
Riaz S, Tenscher AC, Ramming DW, Walker MA (2011) Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor Appl Genet 122:1059–1073
Russell VL (2016) Least-squares means: the R package lsmeans. J Stat Softw 69:1–33. https://doi.org/10.18637/jss.v069.i01
Schaad NW, Postnikova E, Lacy G, Fatmi M, Chang CJ (2004) Xylella fastidiosa subspecies: X. fastidiosa subsp piercei, subsp. nov., X. fastidiosa subsp. multiplex subsp. nov., and X. fastidiosa subsp. pauca subsp. nov. Syst Appl Microbiol 27:290–300
Schuenzel EL, Scally M, Stouthamer R, Nunney L (2005) The multi-gene phylogenetic study of clonal diversity and divergence in North American strains of the plant pathogen Xylella fastidiosa. Appl Environ Microbiol 71:3832–3839
Senthil-Kumar M, Mysore KS (2013) Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol 51:407–427
Simpson AJG, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R et al (2000) Mapping analysis of the Xylella fastidiosa genome. Nature 406:151–159
Stoner WN (1952) A comparison between grape degeneration in Florida and Pierce’s disease in California. Fla Entomol 35:62–68
Stoner WN (1953) Leafhopper transmission of a degeneration of grape in Florida and its relation to Pierce’s disease. Phytopathology 43:611–615
Team RDC (2015) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22:511–519
Thordal-Christensen H (2003) Fresh insights into processes of nonhost resistance. Curr Opin Plant Biol 6:351–357
Van Ooijen J (2006) JoinMap 4, software for the calculation of the genetic linkage maps in experimental populations. Kyazma B. V., Wageningen
Van Ooijen J (2009) MapQTL 6.0: software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B. V., Wageningen
Weising K, Gardner RA (1999) A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42:9–19
Yuan X, Morano L, Bromley R, Spring-Pearson S, Stouthamer R, Nunney L (2010) Multilocus sequence typing of Xylella fastidiosa causing Pierce’s disease and oleander leaf scorch in the United States. Phytopathology 100:601–611
Acknowledgements
Research support from the California Department of Food and Agriculture PD/GWSS Board, and the Louise Rossi Endowed Chair in Viticulture funds is gratefully acknowledged.
Funding
This study was funded by the California Department of Food and Agriculture PD/GWSS Board (Grants nos. 14-0137-SAS and 15-0425-SA).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Reinhard Toepfer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2018_3100_MOESM1_ESM.jpg
Supplementary material 1 Average least square means (lsmeans) comparisons of X. fastidiosa (Xf) levels with Tukey’s test among accessions used as references. Results significantly separate the resistant (b43-17, U505-01, and un-inoculated Chardonnay), intermediate (Roucaneuf and Blanc du Bois) and susceptible (inoculated Chardonnay and U505-22) references that were included in each trial. The use of reference accessions allowed comparison of results across different greenhouse and experiment dates (JPEG 85 kb)
122_2018_3100_MOESM2_ESM.jpg
Supplementary material 2 Phenotypic distribution based on the ELISA bacterial titer (X. fastidiosa (Xf) ln colony forming units/ml) in three breeding populations: a F1 population (n = 47) developed from a cross of the susceptible V. vinifera F2-35 and the resistant accession b41-13, b F1 population (n = 27) developed from a cross of V. vinifera F2-35 and resistant parent ANU67, c F1 population (n = 60) developed with resistant parent T03-16 as female and susceptible V. vinifera cultivar Palomino as male. Arrows indicate the resistant (R) and susceptible (S) parent bacterial count. The number of plants within each titer count are shown on top of the bars (JPEG 64 kb)
Rights and permissions
About this article
Cite this article
Riaz, S., Huerta-Acosta, K., Tenscher, A.C. et al. Genetic characterization of Vitis germplasm collected from the southwestern US and Mexico to expedite Pierce’s disease-resistance breeding. Theor Appl Genet 131, 1589–1602 (2018). https://doi.org/10.1007/s00122-018-3100-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3100-z