Abstract
The diaryl ureas are very important fragments in medicinal chemistry. By means of computer-aided design, 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives were designed and synthesized, and evaluated for their antiproliferative activity against A549, HCT-116, PC-3 cancer cell lines, and HL7702 human normal liver cell lines in vitro by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide colorimetric assay. Most of the target compounds demonstrate significant antiproliferative effects on all the selective cancer cell lines. The calculated IC50 values were reported. The target compound 1-(4-chlorophenyl)-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7u) demonstrated the most potent inhibitory activity (IC50 = 2.39 ± 0.10 μM for A549 and IC50 = 3.90 ± 0.33 μM for HCT-116), comparable to the positive-control sorafenib (IC50 = 2.12 ± 0.18 μM for A549 and IC50 = 2.25 ± 0.71 μM for HCT-116). Conclusively, 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives as the new anticancer agents were discovered, and could be used as the potential BRAF inhibitors for further research.
Similar content being viewed by others
Introduction
Cancer is a major public health problem in developed countries and will become the most serious life-threatening disease worldwide in the near future (Siegel et al. 2013). Some advances in cancer treatment by targeted drugs, such as imatinib, gefitinib, and trastuzumab, were expected to improve cancer cure rates and also to reduce severe adverse actions because of the high specificity of the targeted drugs, which are expressed and have critical roles in cancer cells, but not in normal cells. However, the clinical effect was found to be limited and did not last for a long period because of the acquired resistance of the tumor cells. Furthermore, these drugs often cause on-target and/or off-target severe toxicity (Keefe and Bateman 2011). Therefore, the development of more target-specific therapy, with minimum toxicity, is warranted to extend disease-free survival and improve the quality of life of cancer patients.
Sorafenib is an orally available drug that acts predominantly by targeting the Ras/Raf/MEK (mitogen-activated protein kinase (MAPK)/ERK kinase)/ERK (extracellular signal-regulated kinase) pathway. In vitro sorafenib inhibits several members of the RAF/MEK/ERK signaling cascade, including wild-type BRAF (B-Raf proto-oncogene), mutant BRAF carrying V600E, and serine–threonine kinases CRAF (C-Raf proto-oncogene) (Davies et al. 2002; Moreau et al. 2012; Vultur et al. 2011; Wan et al. 2004; Wilhelm et al. 2004). BRAF is a member of the RAF family of serine–threonine kinases, which are part of the RAS/RAF/MEK/ERK MAPK signaling pathway (Barras 2015; Cheng et al. 2018; Schreck and Rapp 2006). RAF is mainly activated by the G protein, RAS, in response to mutations in cancer cells or by overexpression of different receptor protein tyrosine kinases such as epidermal, platelet derived, and vascular endothelial growth factors (Regad 2015; Sun et al. 2015).
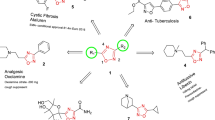
As known, the diaryl urea moiety is widely used in the design of anticancer drugs, such as sorafenib, regorafenib, linifanib, and tivozanib (Fig. 1). And the diaryl ureas are very important fragments in medicinal chemistry and can be used for the synthesis of numerous heterocyclic compounds with diversified biological activities, including antithrombotic (Qiao et al. 2013), antimalarial (Anderson et al. 2013), antibacterial (Keche et al. 2012), and anti-inflammatory (Kulkarni et al. 2013) properties, and they are characterized by their ability to form hydrogen bond interactions with biotargets (Kim et al. 2012; Lu et al. 2014; Xuan et al. 2013). The carbonyl oxygen atom acts as a proton acceptor while the two amide nitrogen atoms are proton donors (Fig. 2). This unique type of structure endows urea derivatives with the ability to bind a variety of enzymes and receptors in the biological systems.
In recent years, proton pump inhibitors (PPIs) as potential anticancer agents were intensively studied in cancer treatment. Lugini et al. compared the antitumor efficacy of different PPIs, including omeprazole, esomeprazole, lansoprazole, rabeprazole, and pantoprazole in vitro and in vivo (Lugini et al. 2016). As shown in Fig. 3, all of the PPIs molecules contain pyridin-2-ylmethyl fragments. It can be predicted that these fragments should play an important role in the antiproliferative activity.
Molecular hybridization strategy is a useful concept in drug design and development based on the combination of pharmacophoric moieties of different bioactive substances to produce a new structure, the affinity and efficacy would be improved, when compared with the parent drugs (Viegas-Junior et al. 2007). These above interesting findings and our continuous quest to identify more potent antiproliferative agents led to the molecular hybridization of diaryl urea and pyridin-2-ylmethyl to integrate them in one molecular platform to generate a new hybrid, and expected that taking this way could get the antiproliferative agents with highly inhibitory activity. The molecular docking results also showed that the urea structure and pyridine can act as a hydrogen bond donor to generate hydrogen bond interactions. Therefore, a novel series of 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives were designed and synthesized.
Results and discussion
Molecular docking studies
A docking study aiming at the characterization of the interaction between the target molecule and BRAF at molecular level was carried out and more-effective BRAF inhibitors were expected to be discovered.
In this study, docking model was selected from Protein Data Bank (PDB) of Research Collaborator for Structural Bioinformatics (RCSB), and the docking study was prepared by Molegro Virtual Docker (MVD) 2008 (v3.0) (Thomsen and Christensen 2006). Initially, the docking studies were carried out on the BRAF—sorafenib complex (PDB code 4R5Y) (Tang et al. 2015), using the Discovery Studio 2016 client to remove the small-molecule ligand of erlotinib in 4R5Y to obtain the three-dimensional crystal structure of BRAF, and then the designed compounds were docked to the crystal structure BRAF with MVD software. The detailed docking data will be described below, using compound 7u with chloro atom in the para position of the phenyl ring, methyl group at the R3 position the trifluoroethoxy group at the R4 position, and H atom at the R5 position on the pyridine ring as an example.
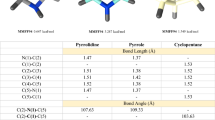
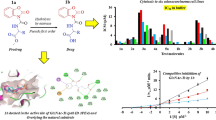
As shown as in the Table 1 and Fig. 4a, it can be seen that compound 7u and sorafenib have similar MolDock scores, it also was inserted inside the protein and has a high degree of overlap in the protein cavity with sorafenib. This implies that the compound we designed is likely to become an effective BRAF inhibitor. From Fig. 4b, c, we can see that the carbon-based oxygen atom in the urea structure in the 7u structure can act as a hydrogen bond acceptor to generate hydrogen bond interaction with the amino-acid residue ASP594. At the same time, NH in the urea structure close to chlorophenyl group can act as a hydrogen bond donor to generate hydrogen bond interactions with the amino-acid residue GLU501. All of this confirms the feasibility of introducing a diphenylurea structure into the drug design to increase the affinity of the compound to the target. Moreover, N atom on the pyridine ring in 7u has a hydrogen bond with THR529, and the N atom on the pyridine with N on amides in sorafenib can generate a hydrogen bond with CYS532. This also shows the importance of introducing a pyridine ring. Hence, we can conclude that both the diaryl urea structure and the pyridinyl ring structure in our designed compounds enhance the affinity of the compound to the target BRAF kinase. In addition, it can be seen that the amino-acid residues HIS574, LYS483, PHE595, VAL471, GLN 530, ALA481, PHE583 formed some weak interactions between the compound 7u and BRAF.
The molecular docking results revealed that the target compound 7u and its analogs were potential inhibitors of BRAF. Therefore, we synthesized a novel series of 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives, aiming to assay their inhibitory activity to BRAF kinase.
Synthesis
The general synthetic route is illustrated in Scheme 1. The reaction of the commercially available 4-nitrophenol (1) with 2-(chloromethyl)pyridine derivatives (2) in ethanol at room temperature obtained compounds 3a–3d (Seto et al. 2005), which converted to key intermediates 4a–4d via Pd-C catalytic hydrogenation reduction (Nguyen et al. 2014). The aryl isocyanates 6a–6f were prepared by reaction between aromatic amines and bis(trichloromethyl)carbonate (BTC) (Liu et al. 2013). Finally, treatment of 4a–4d with arylisocyanates 6a–6f in methylene dichloride yielded 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives (7a–7x) as the target compounds (Liu et al. 2013). The structures of the target compounds and intermediates were characterized by infrared (IR) spectra, proton nuclear magnetic resonance spectra (1H NMR), electrospray ionization mass spectra (ESI-MS), and no target compounds were reported in the literatures.
Bioactivity in cell line
Using sorafenib as a positive control, all of the target compounds were evaluated for the antiproliferative activity in vitro against cancer cell lines, we choose Raf/MEK/ERK pathway cells, including lung cancer A549, and colorectal cancer HCT-116 cell lines, prostate cancer PC-3 cell lines, which Raf/MEK/ERK pathways do not affect (Lee et al. 2005), and human normal liver HL7702 cell lines by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. The antiproliferative assay results evaluated as the half maximal inhibitory concentration (IC50) value (Table 2) and demonstrated that several target compounds have shown moderate to excellent potency against A549 and HCT-116 cancer cell lines, and most of the target compounds hardly exhibit any antiproliferative activities against PC-3 cell lines. Among the target compounds 7u showed the similar inhibitory effect against two cancer cell lines than positive control with IC50 values of 2.39 ± 0.10 and 3.90 ± 0.33 μM, respectively. All the tested compounds exhibit weak cytotoxic activities against HL7702.
All the target compounds could be divided into three classes according to different substituents on the pyridinyl ring (Fig. 5). The analyses of the structure–activity relationships (SARs) were summarized as follows: (1) the results of cell inhibitory activity assay showed, when the 4th position (R4) of the C ring is occupied by the −OCH2CF3 group, the inhibitory activity is optimal. (2) The substituent on the A ring has a significant effect on the inhibitory activity of the target compounds. When the substituents on the C ring are the same, when the ring A of the compound is substituted by an electron withdrawing group such as –F, –Cl or –CF3 or the like, the inhibitory activity is superior to the compound substituted with an electron donating group such as –OCH3. In addition, when electron withdrawing group coexist in the A ring, the inhibitory activity is better than of the two electron withdrawing groups. (3) The inhibitory effect of the compound on the three test cells showed different selectivity. Most of the compounds inhibited the activity of A549 cells better than the inhibitory activities of HCT-116 cells, while the inhibitory activity against PC-3 cells was poor.
Conclusion
Based on the structure of sorafenib, and hybridization, and bioisosterism, combining structural optimization, a novel series of 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives, was designed as the target compounds. The binding mode of 7u, a representative of the target compounds, to BRAF was simulated by MVD software. It was found that NH and oxygen in the urea structure, the N atom of pyridine ring form a hydrogen bond with the amino-acid residue in the BRAF protein. A novel series of 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives were evaluated for their antiproliferative activity against A549, HCT-116, PC-3 cancer cell lines and HL7702 human normal liver cell lines in vitro by using the MTT colorimetric assay. Most of the target compounds demonstrate significant antiproliferative effects on all the selective cancer cell lines. Conclusively, 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives as new anticancer agents were discovered, and could be used as potential lead compounds for further research.
Experimental section
Materials and methods
The reaction process is detected by high performance thin layer chromatography silica gel plate. The melting point was determined by WRX-1S type microscopic melting-point meter, and the thermometer was not corrected. The IR spectra were determined by Bruker IFS55 IR spectrometer (KBr pellets), mass spectrometry data were determined by Waters ACQUITY QDa liquid chromatography-mass spectrometer, and nuclear magnetic resonance spectra were determined by Bruker 400 MHz nuclear magnetic resonance spectrometer with tetramethylsilane as internal standard and perdeuterated dimethyl sulfoxide (DMSO-d6) as solvent.
Preparation of 2-[(4-nitrophenoxy) methyl]pyridine derivatives (3a–3d)
The reaction of the commercially available 4-nitrophenol(0.1 mol) with 2-(chloromethyl)pyridine derivatives (0.1 mol) in ethanol, add 3 mol/L NaOH solution (100 mL) at r.t. and the reaction mixture was stirred for an additional 3 h. After completion of the reaction was monitored TLC, ethanol was removed by vacuum evaporation, the residue was added 200 mL of water, filtered off with suction after 15 min of stirring, placing the filter cake was dried in air, the product was used directly in the next step without purification.
4-(3-methoxypropoxy)-3-methyl-2-[(4-nitrophenoxy)methyl]pyridine (3a)
Yield 90.5% (white solid). ESI-MS (m/z): 333.4 ([M + H]+).
3-methyl-2-[(4-nitrophenoxy)methyl]-4-(2,2,2-trifluoroethoxy)pyridine (3b)
Yield 90.2% (white solid). ESI-MS (m/z): 343.5 ([M + H]+).
4-methoxy-3,5-dimethyl-2-[(4-nitrophenoxy)methyl]pyridine (3c)
Yield 92.5% (white solid). ESI-MS (m/z): 289.0 ([M + H]+).
3,4-dimethoxy-2-[(4-nitrophenoxy)methyl]pyridine (3d)
Yield 95.1% (white solid). ESI-MS (m/z): 291.1 ([M + H]+).
Preparation of 4-(pyridin-2-ylmethoxy) aniline derivatives (4a–4d)
The reaction of the commercially available 2-[(4-nitrophenoxy)methyl]pyridine derivatives (50.0 mmol) with 10% Pd-C in methanol, catalytic hydrogenation reduction reaction at room temperature for 4 h. After completion of the reaction was monitored TLC, palladium carbon was to filter by diatomaceous earth, and then distillation to remove methanol under reduced pressure, the product was used directly in the next step without purification.
4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}aniline (4a)
Yield 90.0% (yellowish oil). ESI-MS (m/z): 329.4 ([M + H]+).
4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}aniline (4b)
Yield 91.7% (yellowish oil). ESI-MS (m/z): 313.1 ([M + H]+).
4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]aniline (4c)
Yield 94.8% (yellowish oil). ESI-MS (m/z): 259.3 ([M + H]+).
4-[(3,4-dimethoxypyridin-2-yl)methoxy]aniline (4d)
Yield 91.1% (yellowish oil). ESI-MS (m/z): 261.2 ([M + H]+).
Preparation of diaryl urea derivatives (7a–7x)
To a solution of BTC (1 mmol) in CH2Cl2 (20 mL) was added dropwise to primary aromatic amine 5 (1 mmol) in CH2Cl2 (20 mL) followed by the dropwise addition of triethylamine (1 mL) in CH2Cl2 (10 mL). The solvent was evaporated. The resulting residue was dissolved in CH2Cl2 (20 mL), and intermediates (6a–6f) (1 mmol) in CH2Cl2 (10 mL) was added dropwise. The mixture was stirred for about 3 h, monitored by TLC. After the reaction completed, the solvent was washed with water and brine, then dried over anhydrous magnesium sulfate. The mixture was filtered, the filtrate was evaporated and purified by silica gel chromatography (CH2Cl2/MeOH = 60/1, v/v) to obtain target compounds.
1-(3,5-difluorophenyl)-3-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}urea (7a)
Yield 56.4% (white solid). m.p.: 126–127 °C. IR (KBr, cm−1): υ 3422.0, 2922.9, 1697.0, 16112, 1554.6, 1514.4, 1478.0, 1459.9, 1430.6, 1407.2, 1383.4, 1291.9, 1233.0, 1203.6, 1151.2, 1111.4, 851.8, 821.6. 1H NMR (400 MHz, DMSO-d6) δ 9.02 (s, 1H), 8.65 (s, 1H), 8.26 (d, J = 5.6 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32 (d, J = 2.2 Hz, 1H), 7.18 (d, J = 2.3 Hz, 1H), 7.16 (d, J = 2.3 Hz, 1H), 7.00 (d, J = 5.7 Hz, 1H), 6.97 (d, J = 2.3 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 6.79–6.73 (m, 1H), 5.11 (s, 2H), 4.11 (t, J = 6.3 Hz, 2H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.95 (m, 2H). ESI-MS (m/z): 458.4 ([M + H]+).
1-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}-3-[3-(trifluoromethyl)phenyl]urea (7b)
Yield 43.6% (white solid). m.p.: 134–135 °C. IR(KBr, cm−1): υ 3424.0, 2922.3, 2852.6, 1600.8, 1560.4, 1511.1, 1384.1, 1339.1, 1296.0, 1164.0, 1117.7, 826.0, 617.0. 1H NMR (400 MHz, DMSO-d6) δ 8.98 (s, 1H), 8.61 (s, 1H), 8.27 (d, J = 5.7 Hz, 1H), 8.01 (s, 1H), 7.56 (d, J = 8.5 Hz, 1H), 7.51–7.48 (m, 1H), 7.36 (d, J = 2.2 Hz, 1H), 7.34 (d, J = 2.3 Hz, 1H), 7.29 (d, J = 7.5 Hz, 1H), 7.00 (d, J = 5.8 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 5.11 (s, 2H), 4.11 (t, J = 6.2 Hz, 2H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.96 (m, 3H). ESI-MS (m/z): 490.3 ([M + H]+); 512.1 ([M + Na]+).
1-(4-chlorophenyl)-3-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}urea (7c)
Yield 49.1% (white solid). m.p.: 158–160 °C. IR(KBr, cm−1): υ 3426.3, 2921.6, 2852.2, 1628.7, 1510.5, 1492.8, 1384.2, 1296.5, 1112.3, 618.3. 1H NMR (400 MHz, DMSO-d6) δ 8.72 (s, 1H), 8.49 (s, 1H), 8.26 (d, J = 5.6 Hz, 1H), 7.47 (d, J = 2.2 Hz, 1H), 7.45 (d, J = 2.2 Hz, 1H), 7.35–7.29 (m, 4H), 7.00 (d, J = 5.7 Hz, 1H), 6.96 (d, J = 2.3 Hz, 1H), 6.95 (d, J = 2.1 Hz, 1H), 5.10 (s, 2H), 4.11 (t, J = 6.2 Hz, 2H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.95 (m, 2H). ESI-MS (m/z): 456.3, 457.2, 458.2, 459.2 ([M + H]+).
1-(4-methoxyphenyl)-3-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}urea (7d)
Yield 47.5% (white solid). m.p.: 145−146 °C. IR(KBr, cm−1): υ 3422.1, 3279.6, 2921.9, 2851.6, 1629.3, 1608.4, 1562.8, 1510.8, 1467.9, 1384.0, 1296.2, 1231.4, 1170.4, 1115.4, 1013.8, 826.1. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (s, 1H), 8.38 (s, 1H), 8.26 (d, J = 5.6 Hz, 1H), 7.34 (d, J = 2.4 Hz, 1H), 7.33 (s, 1H), 7.32 (s, 1H), 7.31 (d, J = 2.2 Hz, 1H), 7.00 (d, J = 5.7 Hz, 1H), 6.95 (s, 1H), 6.93 (s, 1H), 6.86 (d, J = 2.3 Hz, 1H), 6.84 (d, J = 2.2 Hz, 1H), 5.09 (s, 2H), 4.11 (t, J = 6.2 Hz, 2H), 3.71 (s, 3H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.95 (m, 2H). ESI-MS (m/z): 452.3 ([M + H]+).
1-(4-fluorophenyl)-3-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}urea (7e)
Yield 47.9% (white solid). m.p.: 118–119 °C. IR(KBr, cm−1): υ 3422.9, 2922.1, 2852.5, 1643.3, 1562.3, 1508.1, 1463.0, 1384.2, 1294.9, 1210.0, 1118.9, 1045.5, 1014.8, 833.3, 619.5, 521.2. 1H NMR (400 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.48 (s, 1H), 8.27 (d, J = 5.6 Hz, 1H), 7.46–7.44 (m, 1H), 7.44–7.40 (m, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32 (d, J = 2.2 Hz, 1H), 7.11 (d, J = 8.9 Hz, 1H), 7.08 (d, J = 2.2 Hz, 1H), 7.00 (d, J = 5.7 Hz, 1H), 6.96 (d, J = 2.3 Hz, 1H), 6.94 (d, J = 2.1 Hz, 1H), 5.10 (s, 2H), 4.11 (t, J = 6.2 Hz, 2H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.95 (m, 2H). ESI-MS (m/z): 440.2 ([M + H]+).
1-[4-chloro-3-(trifluoromethyl)phenyl]-3-{4-{[4-(3-methoxypropoxy)-3-methylpyridin-2-yl]methoxy}phenyl}urea (7f)
Yield 37.2% (white solid). m.p.: 179–181 °C. IR(KBr, cm−1): υ 3421.6, 2922.7, 1694.9, 1590.8, 1544.7, 1511.5, 1484.5, 1462.9, 1418.4, 1384.0, 1326.7, 1311.7, 1293.2, 1217.1, 1173.4, 1129.3, 1032.4, 826.5, 662.2. 1H NMR (400 MHz, DMSO-d6) δ 9.09 (s, 1H), 8.65 (s, 1H), 8.27 (d, J = 5.6 Hz, 1H), 8.10 (d, J = 2.3 Hz, 1H), 7.64–7.61 (m, 1H), 7.59 (d, J = 8.8 Hz, 1H), 7.36 (d, J = 2.0 Hz, 1H), 7.34 (d, J = 2.3 Hz, 1H), 7.00 (d, J = 5.7 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 5.11 (s, 2H), 4.11 (t, J = 6.2 Hz, 2H), 3.49 (t, J = 6.2 Hz, 2H), 3.25 (s, 3H), 2.18 (s, 3H), 2.02–1.96 (m, 2H). ESI-MS (m/z): 524.3 ([M + H]+).
1-(3,5-difluorophenyl)-3-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}urea (7g)
Yield 46.1% (white solid). m.p.: 162–164 °C. IR(KBr, cm−1): υ 3422.3, 2921.9, 2852.1, 1609.0, 1559.6, 1511.8, 1490.0, 1434.0, 1384.0, 1301.7, 1226.0, 1197.0, 1114.4, 1081.7, 1015.7, 832.6, 672.2. 1H NMR (400 MHz, DMSO-d6) δ 9.03 (s, 1H), 8.65 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 7.35 (d, J = 2.2 Hz, 1H), 7.33 (d, J = 2.2 Hz, 1H), 7.18 (d, J = 2.3 Hz, 1H), 7.16 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 5.5 Hz, 1H), 6.98 (d, J = 2.3 Hz, 1H), 6.96 (d, J = 2.1 Hz, 1H), 6.79–6.73 (m, 1H), 5.04 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 416.3 ([M + H]+); 438.3 ([M + Na]+).
1-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea (7h)
Yield 55.5% (white solid). m.p.: 149–150 °C. IR(KBr, cm−1): υ 3422.1, 2922.2, 2852.1, 1709.7, 1593.6, 1549.4, 1511.4, 1492.4, 1447.2, 1383.7, 1340.2, 1301.0, 1230.8, 1201.7, 1172.2, 1110.6, 1040.4, 818.4, 795.5, 699.6, 666.6, 617.6. 1H NMR (400 MHz, DMSO-d6) δ 8.98 (s, 1H), 8.60 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 8.01 (d, J = 2.1 Hz, 1H), 7.59–7.53 (m, 1H), 7.52–7.48 (m, 1H), 7.36 (d, J = 2.3 Hz, 1H), 7.35 (d, J = 2.1 Hz, 1H), 7.29 (d, J = 7.4 Hz, 1H), 7.15 (d, J = 5.5 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.1 Hz, 1H), 5.04 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 448.3 ([M + H]+); 470.2 ([M + Na]+).
1-(4-chlorophenyl)-3-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}urea (7i)
Yield 42.1% (yellow solid). m.p.: 197–198 °C. IR(KBr, cm−1): υ 3422.2, 2921.8, 2852.4, 1636.1, 1563.1, 1509.5, 1492.6, 1462.8, 1399.6, 1384.3, 1103.3, 825.3, 616.4. 1H NMR (400 MHz, DMSO-d6) δ 8.74 (s, 1H), 8.51 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 7.48 (d, J = 2.2 Hz, 1H), 7.46 (d, J = 2.2 Hz, 1H), 7.35 (d, J = 2.2 Hz, 1H), 7.33 (d, J = 2.1 Hz, 1H), 7.32 (d, J = 2.0 Hz, 1H), 7.30 (d, J = 1.9 Hz, 1H), 7.15 (d, J = 5.5 Hz, 1H), 6.97 (d, J = 2.3 Hz, 1H), 6.95 (d, J = 2.2 Hz, 1H), 5.03 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 414.2, 415.2, 416.2, 417.2 ([M + H]+).
1-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}-3-(4-methoxyphenyl)urea (7j)
Yield 38.9% (white solid). m.p.: 175–176 °C. IR(KBr, cm−1): υ 3422.8, 2922.5, 2852.4, 1599.7, 1561.1, 1509.8, 1384.2, 1299.4, 1226.4, 1110.9, 1037.6, 829.2, 522.9. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (s, 1H), 8.37 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 7.35 (s, 1H), 7.34 (s, 1H), 7.32 (s, 1H), 7.31 (s, 1H), 7.15 (d, J = 5.6 Hz, 1H), 6.95 (d, J = 2.2 Hz, 1H), 6.94 (d, J = 2.2 Hz, 1H), 6.86 (d, J = 2.3 Hz, 1H), 6.84 (d, J = 2.2 Hz, 1H), 5.03 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H), 3.71 (s, 3H). ESI-MS (m/z): 410.3 ([M + H]+); 432.2 ([M + Na]+).
1-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}-3-(4-fluorophenyl)urea (7k)
Yield 48.3% (yellow solid). m.p.: 161–162 °C. IR(KBr, cm−1): υ 3424.9, 2922.8, 2852.8, 1664.8, 1595.7, 1550.6, 1507.0, 1452.7, 1403.4, 1383.8, 1299.9, 1205.9, 1113.4, 1083.5, 1015.5, 832.6, 800.7, 518.3. 1H NMR (400 MHz, DMSO-d6) δ 8.60 (s, 1H), 8.44 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 7.46–7.44 (m, 1H), 7.44–7.41 (m, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32 (d, J = 2.2 Hz, 1H), 7.14 (d, J = 5.5 Hz, 1H), 7.11 (d, J = 8.9 Hz, 1H), 7.08 (d, J = 2.1 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 6.95 (d, J = 2.1 Hz, 1H), 5.03 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 398.2, 399.2 ([M + H]+).
1-[4-chloro-3-(trifluoromethyl)phenyl]-3-{4-[(3,4-dimethoxypyridin-2-yl)methoxy]phenyl}urea (7l)
Yield 40.1% (yellowish solid). m.p.: 106–107 °C. IR(KBr, cm−1): υ 3447.1, 3329.3, 2922.5, 2851.4, 1694.1, 1610.3, 1559.8, 1511.6, 1486.1, 1318.1, 1256.4, 1228.6, 1170.3, 1141.1, 1111.3, 1040.5, 999.7, 874.3, 819.3, 745.2, 686.2, 658.9. 1H NMR (400 MHz, DMSO-d6) δ 9.08 (s, 1H), 8.63 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 8.09 (d, J = 2.4 Hz, 1H), 7.64–7.61 (m, 1H), 7.59 (d, J = 8.8 Hz, 1H), 7.36 (d, J = 2.1 Hz, 1H), 7.34 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 5.6 Hz, 1H), 6.98 (d, J = 2.0 Hz, 1H), 6.96 (d, J = 2.4 Hz, 1H), 5.04 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 482.1, 484.1 ([M + H]+).
1-(3,5-difluorophenyl)-3-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}urea (7m)
Yield 46.6% (white solid). m.p.: 195–196 °C. IR(KBr, cm−1): υ 3421.1, 3298.0, 2922.4, 2852.2, 1642.2, 1606.0, 1563.1, 1510.0, 1477.3, 1444.9, 1383.8, 1292.2, 1234.4, 1202.5, 1159.0, 1118.3, 1102.0, 1002.1, 822.0, 671.3. 1H NMR (400 MHz, DMSO-d6) δ 9.03 (s, 1H), 8.66 (s, 1H), 8.21 (s, 1H), 7.34 (d, J = 2.1 Hz, 1H), 7.33 (d, J = 2.3 Hz, 1H), 7.18 (d, J = 2.3 Hz, 1H), 7.16 (d, J = 2.4 Hz, 1H), 6.99 (d, J = 2.1 Hz, 1H), 6.97 (d, J = 2.3 Hz, 1H), 6.79–6.73 (m, 1H), 5.09 (s, 2H), 3.74 (s, 3H), 2.25 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z): 414.4 ([M + H]+).
1-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}-3-[3-(trifluoromethyl)phenyl]urea (7n)
Yield 41.5% (yellowish solid). m.p.: 158–159 °C. IR(KBr, cm−1): υ 3423.4, 2922.0, 2852.6, 1637.8, 1562.9, 1511.3, 1446.9, 1384.2, 1340.0, 1222.3, 1169.9, 1124.2, 1071.2, 699.6, 617.5. 1H NMR (400 MHz, DMSO-d6) δ 8.99 (s, 1H), 8.62 (s, 1H), 8.22 (s, 1H), 8.01 (d, J = 2.0 Hz, 1H), 7.56 (d, J = 8.5 Hz, 1H), 7.50 (t, J = 7.9 Hz, 1H), 7.36 (d, J = 2.2 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.29 (d, J = 7.6 Hz, 1H), 6.99 (d, J = 2.3 Hz, 1H), 6.97 (d, J = 2.1 Hz, 1H), 5.09 (s, 2H), 3.74 (s, 3H), 2.26 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z):446.3 ([M + H]+); 468.3 ([M + Na]+).
1-(4-chlorophenyl)-3-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}urea (7o)
Yield 48.3% (yellowish solid). m.p.: 202–204 °C. IR(KBr, cm−1): υ 3373.3, 2922.9, 2851.7, 1716.0, 1592.8, 1546.9, 1510.8, 1490.2, 1458.3, 1376.4, 1301.3, 1230.0, 1192.1, 1043.5, 999.8, 828.1, 803.9, 681.3, 507.5. 1H NMR (400 MHz, DMSO-d6) δ 8.76 (s, 1H), 8.53 (s, 1H), 8.22 (s, 1H), 7.48 (d, J = 2.1 Hz, 1H), 7.46 (d, J = 2.2 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.33 (d, J = 2.1 Hz, 1H), 7.32 (d, J = 2.2 Hz, 1H), 7.30 (d, J = 2.1 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 5.09 (s, 2H), 3.74 (s, 3H), 2.25 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z): 412.2, 413.2, 414.2, 415.2 ([M + H]+).
1-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}-3-(4-methoxyphenyl)urea (7p)
Yield 37.4% (white solid). m.p.: 181–182 °C. IR(KBr, cm−1): υ 3420.6, 3286.5, 2923.1, 2852.3, 1632.9, 1597.2, 1557.6, 1503.6, 1464.7, 1383.8, 1297.2, 1259.7, 1219.2, 1170.2, 1106.5, 1032.1, 1003.1, 826.5, 679.8, 560.9. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (s, 1H), 8.38 (s, 1H), 8.22 (s, 1H), 7.34 (s, 1H), 7.33 (s, 1H), 7.32 (s, 1H), 7.31 (s, 1H), 6.96 (d, J = 2.2 Hz, 1H), 6.94 (d, J = 2.2 Hz, 1H), 6.86 (d, J = 2.3 Hz, 1H), 6.84 (d, J = 2.1 Hz, 1H), 5.08 (s, 2H), 3.74 (s, 3H), 3.71 (s, 3H), 2.25 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z): 408.3 ([M + H]+); 430.2 ([M + Na]+).
1-(4-fluorophenyl)-3-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}urea (7q)
Yield 46.4% (white solid). m.p.: 195–196 °C. IR(KBr, cm−1): υ 3425.2, 2922.9, 2852.8, 1638.2, 1607.4, 1561.6, 1508.6, 1042.4, 1384.0, 1293.6, 1262.1, 1215.7, 1106.0, 1004.0, 830.1, 792.1. 1H NMR (400 MHz, DMSO-d6) δ 8.62 (s, 1H), 8.47 (s, 1H), 8.22 (s, 1H), 7.46–7.44 (m, 1H), 7.44–7.41 (m, 1H), 7.34 (d, J = 2.3 Hz, 1H), 7.32 (d, J = 2.1 Hz, 1H), 7.12 (d, J = 5.5 Hz, 1H), 7.09 (d, J = 8.9 Hz, 1H), 6.97 (d, J = 2.3 Hz, 1H), 6.95 (d, J = 2.1 Hz, 1H), 5.08 (s, 2H), 3.74 (s, 3H), 2.25 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z): 396.3 ([M + H]+); 418.2 ([M + Na]+).
1-[4-chloro-3-(trifluoromethyl)phenyl]-3-{4-[(4-methoxy-3,5-dimethylpyridin-2-yl)methoxy]phenyl}urea (7r)
Yield 30.1% (white solid). m.p.: 151–152 °C. IR(KBr, cm−1): υ 3423.6, 2292.3, 2852.1, 1608.9, 1551.8, 1511.7, 1480.6, 1417.3, 1383.9, 1323.7, 1258.1, 1220.0, 1171.8, 1131.9, 1112.3, 1032.8, 1004.9, 823.1, 660.0. 1H NMR (400 MHz, DMSO-d6) δ 9.10 (s, 1H), 8.65 (s, 1H), 8.22 (s, 1H), 8.10 (d, J = 2.3 Hz, 1H), 7.64–7.61 (m, 1H), 7.59 (d, J = 8.8 Hz, 1H), 7.36 (d, J = 2.0 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 6.99 (d, J = 2.0 Hz, 1H), 6.97 (d, J = 2.2 Hz, 1H), 5.10 (s, 2H), 3.74 (s, 3H), 2.26 (s, 3H), 2.22 (s, 3H). ESI-MS (m/z): 480.2 ([M + H]+).
1-(3,5-difluorophenyl)-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7s)
Yield 45.2% (white solid). m.p.: 185–187 °C. IR(KBr, cm−1): υ 3423.3, 2922.2, 2852.5, 2651.0, 1607.4, 1560.6, 1511.2, 1478.5, 1384.3, 1309.9, 1258.1, 1170.2, 1135.0, 1115.9, 1006.6, 834.2, 669.3, 620.0, 580.5. 1H NMR (400 MHz, DMSO-d6) δ 9.07 (s, 1H), 8.69 (s, 1H), 8.35 (d, J = 5.7 Hz, 1H), 7.34 (d, J = 2.1 Hz, 1H), 7.33 (d, J = 2.3 Hz, 1H), 7.18 (d, J = 2.3 Hz, 1H), 7.16 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 5.7 Hz, 1H), 6.98 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 6.78–6.73 (m, 1H), 5.14 (s, 2H), 4.91 (q, J = 8.7 Hz, 2H), 2.21 (s, 3H). ESI-MS (m/z): 468.3 ([M + H]+); 490.2 ([M + Na]+).
1-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}-3-[3-(trifluoromethyl)phenyl]urea (7t)
Yield 61.4% (white solid). m.p.: 162–163 °C. IR(KBr, cm−1): υ 3443.4, 2922.3, 2852.2, 1691.1, 1592.4, 1550.8, 1534.8, 1488.0, 1397.6, 1330.3, 1315.5, 1287.7, 1269.3, 1214.3, 1172.5, 1144.5, 1115.8, 1090.4, 1075.0, 1031.6, 995.3, 916.5, 878.9, 814.8, 747.7, 663.3, 618.1. 1H NMR (400 MHz, DMSO-d6) δ 8.98 (s, 1H), 8.61 (s, 1H), 8.35 (d, J = 5.7 Hz, 1H), 8.00 (s, 1H), 7.55 (d, J = 8.1 Hz, 1H), 7.51–7.48 (m, 1H), 7.36 (s, 1H), 7.34 (d, J = 2.1 Hz, 1H), 7.29 (d, J = 7.6 Hz, 1H), 7.14 (d, J = 5.7 Hz, 1H), 6.99 (d, J = 3.6 Hz, 1H), 6.96 (d, J = 2.0 Hz, 1H), 5.15 (s, 2H), 4.92 (q, J = 8.8 Hz, 2H), 2.21 (s, 3H). ESI-MS (m/z): 500.2 ([M + H]+); 522.1 ([M + Na]+).
1-(4-chlorophenyl)-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7u)
Yield 23.0% (white solid). m.p.: 143–144 °C. IR(KBr, cm−1): υ 4324.2, 2922.0, 2852.7, 1596.1, 1548.9, 1509.9, 1493.0, 1399.6, 1384.1, 1308.7, 1260.4, 1211.0, 1170.2, 1133.5, 828.3, 578.0. 1H NMR (400 MHz, DMSO-d6) δ 8.74 (s, 1H), 8.51 (s, 1H), 8.35 (d, J = 5.6 Hz, 1H), 7.47 (d, J = 2.1 Hz, 1H), 7.46 (d, J = 2.3 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32 (s, 1H), 7.31 (s, 1H), 7.30 (d, J = 2.2 Hz, 1H), 7.14 (d, J = 5.7 Hz, 1H), 6.97 (d, J = 2.1 Hz, 1H), 6.95 (d, J = 2.3 Hz, 1H), 5.14 (s, 2H), 4.92 (q, J = 8.7 Hz, 2H), 2.21 (s, 3H). ESI-MS (m/z): 466.2, 467.2, 468.1, 469.2 ([M + H]+).
1-(4-methoxyphenyl)-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7v)
Yield 19.9% (yellow solid). m.p.: 174–175 °C. IR(KBr, cm−1): υ 3424.7, 2921.8, 2852.5, 1627.5, 1559.2, 1509.3, 1462.0, 1384.2, 1293.8, 1261.3, 1208.2, 1168.8, 1108.7, 828.7, 619.2. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (s, 1H), 8.38 (s, 1H), 8.35 (d, J = 5.6 Hz, 1H), 7.34 (d, J = 2.1 Hz, 1H), 7.33 (d, J = 1.9 Hz, 1H), 7.32 (s, 1H), 7.31 (d, J = 2.1 Hz, 1H), 7.14 (d, J = 5.7 Hz, 1H), 6.95 (d, J = 2.2 Hz, 1H), 6.94 (d, J = 2.2 Hz, 1H), 6.86 (d, J = 2.2 Hz, 1H), 6.84 (d, J = 2.3 Hz, 1H), 5.13 (s, 2H), 4.92 (q, J = 8.7 Hz, 2H), 3.71 (s, 3H), 2.21 (s, 3H). ESI-MS (m/z): 462.2, 463.2 ([M + H]+); 484.2, 485.2 ([M + Na]+).
1-(4-fluorophenyl)-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7w)
Yield 41.4% (yellowish solid). m.p.: 173–175 °C. IR(KBr, cm−1): υ 3425.8, 2922.0, 2852.4, 1630.2, 1561.5, 1508.3, 1402.9, 1384.3, 1261.4, 1206.8, 1170.0, 1108.8, 831.4. 1H NMR (400 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.48 (s, 1H), 8.35 (d, J = 5.7 Hz, 1H), 7.45 (d, J = 5.0 Hz, 1H), 7.43 (d, J = 5.0 Hz, 1H), 7.34 (d, J = 2.2 Hz, 1H), 7.32 (d, J = 2.2 Hz, 1H), 7.14 (d, J = 5.7Hz, 1H), 7.11 (d, J = 8.8 Hz, 1H), 7.08 (d, J = 2.2 Hz, 1H), 6.96 (d, J = 2.2 Hz, 1H), 6.95 (d, J = 2.2 Hz, 1H), 5.14 (s, 2H), 4.92 (q, J = 8.7 Hz, 2H), 2.21 (s, 3H). ESI-MS (m/z): 450.2 ([M + H]+).
1-[4-chloro-3-(trifluoromethyl)phenyl]-3-{4-{[3-methyl-4-(2,2,2-trifluoroethoxy)pyridin-2-yl]methoxy}phenyl}urea (7x)
Yield 21.1% (yellow solid). m.p.: 122–123 °C. IR(KBr, cm−1): υ 3383.8, 2922.6, 2852.7, 1705.5, 1658.6, 1592.0, 1551.4, 1510.9, 1482.0, 1456.7, 1419.2,1383.3, 1259.1, 1172.3, 1133.1, 1032.3, 974.7, 828.2, 664.9. 1H NMR (400 MHz, DMSO-d6) δ 9.08 (s, 1H), 8.63 (s, 1H), 8.22 (d, J = 5.5 Hz, 1H), 8.09 (d, J = 2.4 Hz, 1H), 7.64–7.61 (m, 1H), 7.59 (d, J = 8.8 Hz, 1H), 7.36 (d, J = 2.1 Hz, 1H), 7.34 (d, J = 2.4 Hz, 1H), 7.14 (d, J = 5.6 Hz, 1H), 6.98 (d, J = 2.0 Hz, 1H), 6.96 (d, J = 2.4 Hz, 1H), 5.04 (s, 2H), 3.90 (s, 3H), 3.78 (s, 3H). ESI-MS (m/z): 534.2 ([M + H]+).
Molecular docking
The complex for the enzyme was deposited in the RCSB PDB for the BRAF (PDB code: 4R5Y) (Tang et al. 2015) after eliminating the inhibitor (5-{{(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy}-3,4-dihydro-1,8-naphthyridin-2(1H)-one). The missing residues were built and the polar hydrogen atoms in amino-acid residues were added. Molecular docking was carried out using MVD. The docking was set at 1500 maximum iterations, with a simplex evolution population size of 50 and a minimum of 10 runs. The schematic diagrams of interactions between 4R5Y and the small molecules were analyzed by PyMOL 1.7.
Bioactivity in cell line
The antiproliferative activities of target compounds were determined using a standard MTT assay (Mosmann 1983; Zhang et al. 2017). Exponentially growing cells A549, HCT-116, PC-3, and HL7702 were seeded into 96-well plates and incubated for 24 h to allow the cells to attach. After 24 h of incubation, the culture medium was removed and fresh medium containing various concentrations of the candidate compounds was added to each well. The cells were then incubated for 72 h, thereafter MTT assays were performed and cell viability was assessed at 570 nm by a microplate reader (ThermoFisher Scientific (Shanghai) Instrument Co., Ltd, Shanghai, China). Inhibitory effects were determined as IC50 value.
References
Anderson JW, Sarantakis D, Terpinski J, Kumar TR, Tsai HC, Kuo M, Ager AL, Jacobs Jr WR, Schiehser GA, Ekins S, Sacchettini JC, Jacobus DP, Fidock DA, Freundlich JS (2013) Novel diaryl ureas with efficacy in a mouse model of malaria. Bioorg Med Chem Lett 23:1022–1025
Barras D (2015) BRAF mutation in colorectal cancer: an update. Bio Cancer 7:9–12
Cheng L, Lopez-Beltran A, Massari F, MacLennan GT, Montironi R (2018) Molecular testing for BRAF mutations to inform melanoma treatment decisions: a move toward precision medicine. Mod Pathol 31:24–38
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in human cancer. Nature 417:949–954
Keche AP, Hatnapure GD, Tale RH, Rodge AH, Birajdar SS, Kamble VM (2012) A novel pyrimidine derivatives with aryl urea, thiourea and sulfonamide moieties, synthesis, anti-inflammatory and antimicrobial evaluation. Bioorg Med Chem Lett 22:3445–3448
Keefe DM, Bateman EH (2011) Tumor control versus adverse events with targeted anticancer therapies. Nat Rev Clin Oncol 9:98–109
Kim HJ, Cho HJ, Kim H, El-Gamal MI, Oh CH, Lee SH, Sim T, Hah JM, Yoo KH (2012) New diarylureas and diarylamides possessing acet(benz)amidophenyl scaffold, design, synthesis, and antiproliferative activity against melanoma cell line. Bioorg Med Chem Lett 22:3269–3273
Kulkarni RG, Laufer S, Mangannavar C, Garlapati A (2013) Design, synthesis and characterization of N’,N”-diaryl ureas as p38 kinase inhibitors. Med Chem 9:213–221
Lee Jr JT, Steelman LS, McCubrey JA (2005) Modulation of Raf/MEK/ERK kinase activity does not affect the chemoresistance profile of advanced prostate cancer cells. Int J Oncol 26:1637–1644
Liu DZ, Tian Z, Yan ZH, WuL X, Ma Y, Wang Q, Liu W, Zhou HG, Yang C (2013) Design, synthesis and evaluation of 1,2-benzisothiazol-3-one derivatives as potent caspase-3 inhibitors. Bioorg Med Chem 21:2960–2967
Lu C, Tang K, Li Y, Li P, Lin Z, Yin D, Chen X, Huang H (2014) Design, synthesis and evaluation of novel diaryl urea derivative as potential antitumor agents. Eur J Med Chem 77:351–360
Lugini L, Federici C, Borghi M, Azzarito T, Marino ML, Cesolini A, Spugnini EP, Fais S (2016) Proton pump inhibitors while belonging to the same family of generic drugs show different anti-tumor effect. J Enzyme Inhib Med Chem 31:538–545
Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Helias-Rodzewicz Z, Marin C, Peschaud F, Chagnon S, Zimmermann U, Clerici T, Emile J (2012) Prognostic value of BRAF V600 mutations in melanoma patients after resection of metastatic lymph nodes. Ann Surg Oncol 19:4314–4321
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Nguyen T, Yang TM, Go ML (2014) Functionalized acridin-9-yl phenylamines protected neuronal HT22 cellsfrom glutamate-induced cell death by reducing intracellular levels of free radical species. Bioorg Med Chem Lett 24:1830–1838
Qiao JX, Wang TC, Ruel R, Thibeault C, L’Heureux A, Schumacher WA, Spronk SA, Hiebert S, Bouthillier G, Lloyd J, Pi Z, Schnur DM, Abell LM, Hua J, Price LA, Liu E, Wu Q, Steinbacher TE, Bostwick JS, Chang M, Zheng J, Gao Q, Ma B, McDonnell PA, Huang CS, Rehfuss R, Wexler RR, Lam PY (2013) Conformationally constrained ortho-anilino diaryl ureas: discovery of 1-(2-(1’-neopentylspiro[indoline-3,4’-piperidine]-1-yl)phenyl)-3-(4-(trifluoromethoxy)phenyl)urea, a potent, selective, and bioavailable. P2Y1 J Med Chem 56:9275–9295
Regad T (2015) Targeting RTK signaling pathways in cancer. Cancers 7:1758–1784
Schreck R, Rapp UR (2006) Raf kinases: oncogenesis and drug discovery. Int J Cancer 119:2261–2271
Seto M, Miyamoto N, AikawaK, Aramaki Y, Kanzaki N, Iizawa Y, Baba M, Shiraishia M (2005) Orallyactive CCR5 antagonists as anti-HIV-1 agents. Part 3: synthesis and biological activities of 1-benzazepine derivatives containing a sulfoxide moiety. Bioorg Med Chem 13:363–386
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30
Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF (2015) Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res 35:600–604
Tang Z, Yuan X, Du R, Cheung SH, Zhang G, Wei J, Zhao Y, Feng Y, Peng H, Zhang Y, Du Y, Hu X, Gong W, Liu Y, Gao Y, Liu Y, Hao R, Li S, Wang S, Ji J, Zhang L, Li S, Sutton D, Wei M, Zhou C, Wang L, Luo L (2015) BGB-283, a novel RAF kinase and EGFR Inhibitor, displays potent antitumor activity in BRAF-mutated colorectal cancers. Mol Cancer Ther 14:2187–2197
Thomsen R, Christensen MH (2006) MolDock: a new technique for high-accuracy molecular docking. J Med Chem 49:3315–3321
Viegas-Junior C, Danuello A, da Silva Bolzani V, Barreiro EJ, Fraga CA (2007) Molecular hybridization: a useful tool in the design of new drug prototypes. Curr Med Chem 14:1829–1852
Vultur A, Villanueva J, Herlyn M (2011) Targeting BRAF in advanced melanoma: a first step toward manageable disease. Clin Cancer Res 17:1658–1663
Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Project CG, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R (2004) Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116:855–867
Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, Cao Y, Shujath J, Gawlak S, Eveleigh D, Rowley B, Liu L, Adnane L, Lynch M, Auclair D, Taylor I, Gedrich R, Voznesensky A, Riedl B, Post LE, Bollag G, Trail PA (2004) BAY 43-9006 exhibits broad spectrum oral antitum or activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res 64:7099–7109
Xuan W, Ding W, Hui HX, Zhang SQ (2013) Synthesis and cytotoxic activity of diaryl urea derivatives with a 4-methylpiperazinylcarbonyl moiety. Med Chem Res 22:3857–3862
Zhang L, Deng XS, Zhang C, Meng GP, Wu JF, Li XS, Zhao QC, Hu C (2017) Design, synthesis and cytotoxic evaluation of a novel series of benzo[d]thiazole-2-carboxamide derivatives as potential EGFR inhibitors. Med Chem Res 26:2180–2189
Acknowledgements
This work was supported by the National Science Foundation of China (NSFC) for the grant No. 21342006 and the Program for Innovative Research Team of the Ministry of Education of China for the grant No. IRT_14R36. The authors would like to thank Molegro ApS for kindly providing a free evaluation copy of their software package and Schrödinger, LLC for allowing the use of PyMOL Molecular Graphics System.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Feng, J., Li, T., Liang, S. et al. Synthesis and biological evaluation of a new series of 1-aryl-3-[4-(pyridin-2-ylmethoxy)phenyl]urea derivatives as new anticancer agents. Med Chem Res 29, 1413–1423 (2020). https://doi.org/10.1007/s00044-020-02554-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-020-02554-z