Abstract
The endogenous mitochondrial quality control (MQC) system serves to protect mitochondria against cellular stressors. Although mitochondrial dysfunction contributes to cardiac damage during many pathological conditions, the regulatory signals influencing MQC disruption during septic cardiomyopathy (SC) remain unclear. This study aimed to investigate the involvement of pyruvate kinase M2 (PKM2) and prohibitin 2 (PHB2) interaction followed by MQC impairment in the pathogenesis of SC. We utilized LPS-induced SC models in PKM2 transgenic (PKM2TG) mice, PHB2S91D-knockin mice, and PKM2-overexpressing HL-1 cardiomyocytes. After LPS-induced SC, cardiac PKM2 expression was significantly downregulated in wild-type mice, whereas PKM2 overexpression in vivo sustained heart function, suppressed myocardial inflammation, and attenuated cardiomyocyte death. PKM2 overexpression relieved sepsis-related mitochondrial damage via MQC normalization, evidenced by balanced mitochondrial fission/fusion, activated mitophagy, restored mitochondrial biogenesis, and inhibited mitochondrial unfolded protein response. Docking simulations, co-IP, and domain deletion mutant protein transfection experiments showed that PKM2 phosphorylates PHB2 at Ser91, preventing LPS-mediated PHB2 degradation. Additionally, the A domain of PKM2 and the PHB domain of PHB2 are required for PKM2-PHB2 binding and PHB2 phosphorylation. After LPS exposure, expression of a phosphorylation-defective PHB2S91A mutant negated the protective effects of PKM2 overexpression. Moreover, knockin mice expressing a phosphorylation-mimetic PHB2S91D mutant showed improved heart function, reduced inflammation, and preserved mitochondrial function following sepsis induction. Abundant PKM2 expression is a prerequisite to sustain PKM2-PHB2 interaction which is a key element for preservation of PHB2 phosphorylation and MQC, presenting novel interventive targets for the treatment of septic cardiomyopathy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a severe complication of sepsis, septic cardiomyopathy (SC) manifests as reversible myocardial depression in patients with septic shock. Clinical features of SC include left ventricular dilation, decreased myocardial contractile function, and increased end-diastolic ventricular volume. Inflammatory injury, aberrant immune response, and cytokine storm have been proposed as the molecular bases of decreased myocardial function during sepsis-induced cardiomyopathy [1, 2]. However, findings from a randomized study in patients with virus-negative inflammatory cardiomyopathy suggest that immunosuppressive strategies may not provide additional clinical benefits for patients with SC [3, 4]. A growing body of evidence highlights the functional significance of mitochondrial damage in the pathogenesis of SC. Increased mitochondrial fission compromises cardiomyocyte energy status, leading to impaired myocardial contraction [5]. Additionally, defective mitophagy contributes to oxidative stress and thus mitochondria-dependent apoptosis in the myocardium [6, 7]. Decreased mitochondrial metabolism, reflected by impaired mitochondrial respiration, has been found to be closely associated with myocardial swelling and inflammation [8]. Such evidence suggests multiple mechanisms linking mitochondrial dysfunction to the pathogenesis of SC. However, the upstream mediators and signal transduction mechanisms responsible for inducing mitochondrial abnormalities in cardiomyocytes during sepsis remain incompletely characterized.
Mitochondrial quality control (MQC) is an inherent protective mechanism that regulates mitochondrial dynamics, mitophagy, biogenesis, and the mitochondrial unfolded protein response (mtUPR) to maintain normal mitochondrial morphology and function [9,10,11,12,13]. Our previous studies have reported the beneficial effects of MQC in the setting of cardiac ischemia-reperfusion injury (IRI) [14, 15]. In IRI-challenged cardiac cells with preserved MQC, mitochondrial potential was maintained, mitochondrial oxidative stress was neutralized, mitochondrial permeability transition pore (mPTP) opening was inhibited, and mitochondria-induced apoptosis was suppressed [14]. Such a scenario is supported by a recent study in a mouse model of type-2 diabetes [16], which delineated the cardioprotective impact of MQC on diabetic cardiomyopathy via upregulating mitochondrial respiration, accelerating mitochondrial ATP production, and reducing mitochondrial ROS leakage. By comparison, defective MQC is regarded as a contributory factor augmenting vanadium-mediated cardiovascular toxicity, possibly through inducing oxidative stress, mitochondrial vacuolar degeneration, and cardiomyocyte apoptosis [2, 17]. Despite this evidence, the specific alterations in MQC occurring during SC, as well as their underlying mechanisms, have not been entirely clarified.
Prohibitin 2 (PHB2) assembles at the inner mitochondrial membrane to form a supra-macromolecular structure that serves to maintain mitochondrial integrity, fusion/fission balance [18], biogenesis [19], and mitophagy [20, 21]. Evidence strongly indicates that PHB2 plays a pivotal role in coordinating the mitochondrial response to various stressors by regulating crucial aspects such as mitochondrial redox biology, mtDNA stability, membrane integrity, and respiratory activity [20,21,22,23]. While these results suggest that PHB2 functions as an essential MQC mediator, the impact of potential changes in PHB2 expression or stability in SC has not been determined.
Pyruvate kinase M2 (PKM2), one of the rate-limiting enzymes of glycolysis, critically affects various aspects of mitochondrial bioenergetics. PKM2 assembles in dimeric and tetrameric conformations. Tetrameric PKM2 has pyruvate kinase (PK) activity and catalyzes the production of pyruvate by phosphoenolpyruvate (PEP) [24]. Dimeric PKM2 has lower PK activity and exerts also non-glycolytic enzymatic functions, acting for instance as a transcription factor and a protein kinase [24]. Importantly, PKM2 may show ample intracellular distribution, e.g. attached to mitochondria, to the endoplasmic reticulum, and translocated to the nucleus [25]. Although mitochondrial dynamics [26], mitophagy [27], and biogenesis [28] have been reported to be controlled by PKM2, such influence has not been evaluated in the setting of SC. Considering the beneficial effects offered by PKM2 overexpression on heart failure [29] and myocardial infarction [30], we asked whether SC may be alleviated by PKM2 through stabilizing PHB2 and supporting MQC in cardiomyocytes.
Results
Downregulated PKM2 expression contributes to myocardial injury in SC
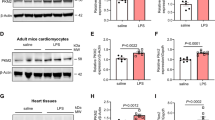
To assess potential alterations in PKM2 expression during SC, qPCR and western blots were performed using heart tissues from LPS-treated mice. Both PKM2 mRNA transcription (Fig. 1A) and protein expression (Fig. 1B and C) were significantly downregulated compared to control tissues from WT mice. Importantly, upon LPS treatment, the levels of tetrameric PKM2 in cardiac tissue of WT mice remained unchanged, whereas a significant reduction was observed in the levels of dimeric PKM2 (Fig. 1D and E). This suggests that LPS mainly suppresses the content of dimeric PKM2 in the heart. To evaluate the potential cause-effect of PKM2 downregulation on myocardial injury during SC, we generated PKM2 transgenic (PKM2Tg) mice [31]. PKM2Tg mice were born and developed normally with no detectable pathological symptoms, and showed ordinary phenotypes, lifespan, body weight, and heart function until 24 months of age. Western blots showed that cardiac levels of total PKM2 were similar, while those of tetrameric and dimeric PKM2 were slightly increased, in PKM2Tg compared to WT mice (Fig. 1D and E).
PKM2 downregulation contributes to myocardial injury in SC. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. (A) Analysis of transcriptional levels of cardiac Pkm2 in WT mice by qPCR. (B-E) Western blot analysis of total PKM2, dimeric PKM2, and tetrameric PKM2 protein expression in mouse heart tissues. (F) Survival analysis of control and PKM-overexpressing mice after SC induction. (G-M) Assessment of cardiac function in WT or PKM2Tg mice by echocardiography. LVDd, left ventricular diastolic dimension; LVDs, left ventricular systolic dimension; IVS, interventricular septum thickness; E/A, ratio of early to late transmitral flow velocities; FS, ratio of left ventricular fractional shortening. (N) HE staining of myocardial tissue. (O) Representative images of electron microscopy of heart tissues. Yellow arrows indicate disorganized muscle fibers and deformed mitochondria with edematous cristae. (P-R) ELISA-based analysis of LDH, TnI, and CK-MB levels in mice sera. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
Compared to WT mice, the mortality of PKM2Tg mice was significantly reduced after SC induction (Fig. 1F). Moreover, upon LPS challenge, cardiac function assessed by echocardiogram, was impaired in WT mice and largely preserved in PKM2Tg mice (Fig. 1G and M). Specifically, compared with WT mice, PKM2 overexpression markedly improved cardiac systolic function, as evidenced by normalized LVEF, FS, and LVSd after LPS administration (Fig. 1G and I). Similarly, alterations related to ventricular dilation were also ameliorated in PKM2Tg mice compared with WT mice (Fig. 1J and M). To assess the presence of cardiac functional abnormalities associated with myocardial structural disorder, HE staining and electron microscopy (EM) were employed to detect myocardial morphology and cardiomyocyte ultrastructure. As expected, histological analysis indicated myocardial swelling in WT heart tissues accompanied by disorganized muscle fibers, vacuolated cardiomyocytes, and deformed mitochondria with edematous cristae (Fig. 1N and O). However, these structural derangements were not observed in LPS-challenged PKM2Tg mice (Fig. 1N and O).
Myocardial injury is also featured by increased levels of myocardial damage biomarkers such as TnI, CK-MB, and LDH. After exposure to LPS, the concentrations of TnI, CK-MB, and LDH in sera were rapidly upregulated in WT mice and drastically relieved in PKM2Tg mice (Fig. 1P and R). These results highlight a cardioprotective role of PKM2 in the setting of SC.
PKM2 overexpression reduces myocardial inflammation in SC
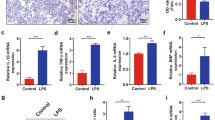
Abnormal inflammation has been identified as a core pathological factor aggravating myocardial damage under septic conditions. Thus, we sought to investigate whether PKM2 overexpression is capable to attenuate myocardial inflammatory reactions during sepsis. ELISA assay revealed a significant upregulation of IL-6 and CRP concentrations in the serum of WT mice following LPS treatment, whereas LPS-treated PKM2Tg mice exhibited reduced levels of IL-6 and CRP (Fig. 2A and B). Besides, the transcriptions of pro-inflammation factors, such as Mmp9, Mcp1 and Tnfα, were obviously elevated in WT heart tissue but maintained at near normal levels in PKM2-overexpressed mice after LPS treatment (Fig. 2C and E). Parallel increased pro-inflammatory cytokines and factors, adhesion proteins, i.e. ICAM1 (Fig. 2F and G) and VCAM1 (Fig. 2H and I), was also observed in heart tissues in WT mice but only remarkably recovered in PKM2Tg mice. Accordingly, immunofluorescence showed that Gr-1-positive neutrophils were recruited into the myocardium of LPS-treated WT, but not PKM2Tg, mice (Fig. 2J and K). These findings indicate that PKM2 overexpression elicits an anti-inflammatory action in SC.
PKM2 overexpression reduces LPS-mediated myocardial inflammation. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. (A, B) ELISA-based measurement of IL-6 and CRP levels in mouse serum. (C-E) Transcriptional analysis of cardiac Mmp9, Mcp1, and Tnfα expression in WT or PKM2Tg mice by qPCR. (F, G) Quantification and representative images of immunohistochemical staining of ICAM1 in heart tissues. (H, I) Quantification and representative images of immunohistochemical staining of VCAM1 in heart tissues. (J, K) Immunofluorescent staining of Gr-1-positive neutrophils in heart tissues. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
PKM2 overexpression preserves cardiomyocyte viability during SC
Cardiomyocyte dysfunction and death are central events contributing to decreased myocardial function during SC. To assess whether enhanced PKM2 expression promotes cardiomyocyte viability after LPS exposure, mouse cardiac muscle HL-1 cells transfected with PKM2-overexpressing adenovirus (Ad-PKM2) or β-gal adenovirus (Ad-β-gal; control) were subjected to LPS to simulate SC in vitro. CCK-8 assays showed that LPS reduced cellular viability in cells transfected with Ad-β-gal rather than with Ad-PKM2 (Fig. 3A). Besides, TUNEL staining showed that LPS promoted apoptosis in Ad-β-gal-expressing cardiomyocytes, while this alteration was reversed in cells transfected with Ad-PKM2 (Fig. 3B and C). Accordingly, ELISA demonstrated that following LPS treatment cardiac caspase-3 activity was augmented in WT mice but remained at near-normal levels in PKM2Tg mice (Fig. 3D). TUNEL assays further showed that LPS promoted extensive cardiomyocyte death in WT mice, and this effect was significantly attenuated in PKM2Tg mice (Fig. 3E and F).
PKM2 overexpression preserves cardiomyocyte viability after septic insult. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo and ex-vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. Immortalized mouse cardiac muscle HL-1 cells were treated with 10 µg/mL of LPS for 24 h to simulate SC in vitro. Cells treated with an equal volume of phosphate buffer saline were used as controls. Before LPS treatment, cardiomyocytes were transduced with adenovirus encoding PKM2 (Ad-PKM2). β-gal-overexpressing (Ad-β-gal) cells were used as controls. (A) Cell viability was measured in vitro via CCK-8 assay. (B, C) TUNEL staining was applied to analyze the number of apoptotic HL-1 cells in vitro in the presence of LPS. (D) ELISA-based measurement of cardiac caspase-3 activity following SC induction in vivo. (E, F) Results of TUNEL assays conducted in heart tissues following SC induction in vivo. (G-L) Analysis of contraction and relaxation parameters in field-stimulated single adult cardiomyocytes isolated from WT and PKM2Tg mice. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
To assess whether PKM2 overexpression would also normalize cardiomyocyte function in the setting of SC, contraction/relaxation properties were measured in primary cardiomyocytes isolated from WT and PKM2Tg mice after being subjected to electrical field stimulation. LPS treatment had no impact on resting adult cardiomyocyte length in any mice type (Fig. 3G). However, contractile parameters, including peak shortening (PS), maximal velocity of shortening (+ dL/dt), and time-to-peak shortening (TPS) (Fig. 3H and J), as well as relaxation parameters, i.e. maximal velocity of relengthening (-dL/dt) and time to 90% relengthening (TR90) (Fig. 3K and L), were impaired in LPS-treated WT cardiomyocytes. In contrast, contraction/relaxation properties were largely preserved in LPS-challenged PKM2Tg cardiomyocytes. These data indicate that PKM2 supports cardiomyocyte viability and mechanical function in the setting of SC.
PKM2 protects mitochondrial function in LPS-challenged cardiomyocytes
Mitochondrial damage critically contributes to cardiomyocyte dysfunction and death caused by SC [32]. Based on the above findings, we proceeded to investigate whether PKM2 overexpression was able to prevent or attenuate LPS-mediated mitochondrial damage in cultured cardiomyocytes. Initially, we utilized the mtDNA/nDNA ratio to assess potential changes in mitochondrial content or copy numbers in response to PKM2 overexpression both in vivo and in vitro. Notably, the mtDNA/nDNA ratio remained unchanged in PKM2Tg mice compared to WT mice (Fig. 4A). Additionally, Ad-PKM2 transfection had no impact on the mtDNA/nDNA ratio (Fig. 4B). Further, as depicted in Fig. 4C and D, mitochondrial membrane potential was reduced in Ad-β-gal-transduced, control HL-1 cells, but remained stable in those overexpressing PKM2 after LPS exposure. Besides, mitochondrial ROS production was enhanced by LPS in control cells, whereas this effect was attenuated by Ad-PKM2 overexpression (Fig. 4E and F). Moreover, mitochondrial DNA (mtDNA) copy number (Fig. 4G) and mtDNA translation (Fig. 4H) were impaired in response to LPS in control cells, but largely preserved instead in PKM2-overexpressing cells. To further evaluate mitochondrial function in vitro, Seahorse assay was performed. Analysis of mitochondrial oxygen consumption rate (OCR) revealed that LPS treatment significantly impaired mitochondrial respiration in control cardiomyocytes, whereas PKM2-overexpressing cardiomyocytes were not affected (Fig. 4I and M). Collectively, our findings confirm that PKM2 overexpression can alleviate LPS-induced mitochondrial dysfunction in cardiac cells.
PKM2 protects mitochondrial function in cardiomyocytes exposed to LPS. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. Immortalized mouse cardiac muscle HL-1 cells were treated with 10 µg/mL of LPS for 24 h to simulate SC in vitro. Cells treated with an equal volume of phosphate buffer saline were used as controls. Before LPS treatment, cardiomyocytes were transduced with Adenovirus encoding PKM2 (Ad-PKM2). β-gal-overexpressing (Ad-β-gal) cells were used as controls. (A) The CO1 gene of mtDNA and the NDUFV1 gene of nDNA were amplified using qPCR to assess the relative ratio of mtDNA/nDNA in heart tissues obtained from WT or PKM2Tg mice under normal physiological conditions. (B) HL-1 cells were transfected with Ad-PKM2 or Ad-β-gal. Subsequently, the CO1 gene of mtDNA and the NDUFV1 gene of nDNA were amplified using qPCR to evaluate the relative mtDNA/nDNA ratio in vitro. (C, D) Analysis of the effect of LPS exposure on mitochondrial membrane potential in JC-1-loaded HL-1 cells overexpressing adenovirus PKM2 (Ad-PKM2) or β-gal (Ad-β-gal; control). (E, F) Detection of mitochondrial ROS production in HL-1 cells loaded with MitoSOX Red. (G, H) Estimation of mitochondrial DNA (mtDNA) copy number and transcription via qPCR. (I-M) Mitochondrial oxygen consumption rate (OCR) was determined by the Seahorse XF realtime ATP rate assay using an XF-24 Extracellular Flux Analyzer. ATP turnover, baseline OCR, maximal respiration capacity and proton leak were measured in HL-1 cells treated with Ad-β-gal or Ad-PKM2. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
PKM2 overexpression maintains mitochondrial quality control in cardiomyocytes challenged with LPS
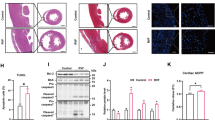
In response to mitochondrial damage, a series of endogenous adaptive mechanisms, including mitochondrial fission/fusion, mitophagy, organelle biogenesis, and the mitochondrial unfolded protein response (mtUPR), collectively exerting mitochondrial quality control (MQC), are engaged to preserve mitochondrial morphology and function [15, 33, 34]. To study the molecular basis underlying the protective effect of PKM2 expression on mitochondrial integrity in cardiomyocytes challenged with LPS, we investigated whether MQC was governed and/or normalized by PKM2. Quantitative protein analysis showed that LPS exposure upregulated levels of Drp1 and Fis1 and suppressed the expression of Mfn2 and Opa1 (Fig. 5A and E), indicative of increased fission and decreased fusion in WT mouse heart tissues. Whereas these changes were negated by PKM2 overexpression. In vitro immunofluorescence analysis of HL-1 cardiomyocyte mitochondria showed that the mitochondrial network was disrupted in the presence of LPS (Fig. 5F and H), with a significant drop in average mitochondrial length and an elevation in the proportion of cardiomyocytes with fragmented mitochondria. In contrast, mitochondrial morphology was preserved in cardiomyocytes overexpressing PKM2 (Fig. 5F and H).
PKM2 overexpression maintains mitochondrial quality control. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. Immortalized mouse cardiac muscle HL-1 cells were treated with 10 µg/mL of LPS for 24 h to simulate SC in vitro. Cells treated with an equal volume of phosphate buffer saline were used as controls. Before LPS treatment, cardiomyocytes were transduced with Adenovirus encoding PKM2 (Ad-PKM2). β-gal-overexpressing (Ad-β-gal) cells were used as controls. (A-E). Western blot analysis of cardiac Drp1, Fis1, Mfn2, and Opa1 expression in vivo. (F-H) Results of immunofluorescence analysis of mitochondrial morphology in HL-1 cells. Rate of cells with fragmented mitochondria (F), average length of mitochondria (G), and representative mitochondrial immunofluorescence images (H) are shown. (I-M) Representative immunoblots and quantification of changes in Parkin, Atg5, Beclin1, and PGC1α expression in cardiac tissues. (N, O). Representative images and quantitative results of mt-Keima assays assessing mitophagy in HL-1 cells. (P-T) Transcriptional analysis of cardiac Nrf2, Tfam, Atf6, LonP1, and mtHsp70 expression by qPCR in vivo. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
Regarding mitophagy and mitochondrial biogenesis, western blot analysis revealed that cardiac levels of Parkin, Atg5, Beclin1, and PGC1α were significantly downregulated in WT mice after LPS treatment. In contrast, PKM2Tg mice exhibited near-normal levels of these proteins (Fig. 5I and M). Mt-Keima assays, used to monitor mitophagy activity, showed in turn that mitophagic flux was impaired by LPS in control HL-1 cells and restored after PKM2 overexpression (Fig. 5N and O). In addition, the transcriptions of Nrf2 and Tfam, two downstream effectors of PGC1α-mediated mitochondrial biogenesis, were blunted by LPS in WT mice but restored upon PKM2 overexpression (Fig. 5P and Q). The mtUPR, encompasses an adaptive transcriptional stress program, activated by disrupted mitochondrial proteostasis, that mediates the expression of genes in response to stress [35]. We noted that after SC induction, cardiac transcriptions of Atf6, LonP1, and mtHsp70 were upregulated in WT mice but maintained to low levels in PKM2Tg mice comparable with untreated WT mice (Fig. 5R and T). Thus, the above data showed that PKM2 overexpression maintains MQC function in cardiac cells exposed to LPS.
PKM2 binds to and prevents PHB2 degradation
Recent research has identified PHB2 as a regulator of MQC [36]. To discern the molecular mechanism by which PKM2 preserves MQC in LPS-induced SC, we focused on potential alterations in PHB2 expression. qPCR analysis showed that neither LPS nor PKM2 overexpression influenced cardiac PHB2 mRNA expression (Fig. 6A). However, western blot assays showed that after LPS administration cardiac PHB2 expression was downregulated in WT mice and sustained at near-baseline levels in PKM2Tg mice (Fig. 6B and C). The correlation between PHB2 and PKM2 expression led us to speculate that the post- translational stability of PHB2 may be under the control of PKM2. Pulse-chase analysis on HL-1 cells further showed that LPS increased the degradation rate of endogenous PHB2 protein in control cells, and this effect was prevented by PKM2 overexpression (Fig. 6D and E).
PKM2 binds to and prevents PHB2 degradation. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. Immortalized mouse cardiac muscle HL-1 cells were treated with 10 µg/mL of LPS for 24 h to simulate SC in vitro. Cells treated with an equal volume of phosphate buffer saline were used as controls. Before LPS treatment, cardiomyocytes were transduced with Adenovirus encoding PKM2 (Ad-PKM2). β-gal-overexpressing (Ad-β-gal) cells were used as controls. (A) Analysis of the effect of LPS exposure on cardiac Phb2 mRNA expression levels in WT and PKM2Tg mice. (B, C) Western blots analysis of cardiac PHB2 protein levels in WT and PKM2Tg mice. (D, E) Analysis of the half-life of PHB2 protein in HL-1 cells (pulse-chase assay). (F-G) Western blot analysis of PHB2 expression in HL-1 cells exposed to LPS in the presence or absence of MG132, a proteasome inhibitor, or betulinic acid (BA), a proteasome activator. (H, I) Pulse-chase analysis of PHB2 degradation rate in cultured in HL-1 cells treated with LPS and MG132. (J, K) Pulse-chase analysis of PHB2 degradation rate in cultured in HL-1 cells treated with LPS and BA. (L-M) Mapping of PHB2/PKM2 interacting regions by docking analysis. (N, O) Co-IP analysis of PKM2/PHB2 binding in HL-1 cells transfected with different domain deletion PKM2 mutants. (P) Mapping of PKM2 regions and deletion mutants. (Q) Western blot analysis of PHB2 expression in HL-1 cells transfected with domain deletion PKM2 mutants. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05
In line with these findings, LPS-mediated PHB2 downregulation was nullified upon incubation of HL-1 cardiomyocytes with the proteasome inhibitor MG132 (Fig. 6F-G), suggesting involvement of the ubiquitin-proteasome system. On the contrary, and confirming proteasome involvement, PKM2 overexpression failed to sustain PHB2 expression in LPS-treated cardiomyocytes pre-incubated with betulinic acid (BA), a potent proteasome activator (Fig. 6H-I). Accordingly, protein stability analysis using pulse-chase assays showed that MG132 exposure counteracted LPS-stimulated PHB2 degradation (Fig. 6J), whereas PKM2 overexpression failed to extend the half-life of PHB2 in BA-treated cardiomyocytes (Fig. 6K). These results suggested that PKM2 overexpression in cardiomyocytes attenuates proteasome-mediated PHB2 degradation.
To elucidate the molecular basis underlying PKM2-mediated PHB2 stability, we evaluated the presence of constitutive interactions between PKM2 and PHB2 in cultured HL-1 cells. First, the active regions of PKM2 and PHB2 were screened by molecular docking simulations. We detected the existence of H-bond or hydrophobic interactions between PKM2 and PHB2 (Fig. 6L and M), with a minimum binding energy of -12.3 kcal·mol− 1. Co-IP assays further confirmed an endogenous interaction between PKM2 and PHB2 under physiological condition or LPS stress (Fig. 6N and O). To define the structural moieties required for PKM2/PHB2 binding, we first mapped the regions of PKM2 that are required for protein-protein binding. Since PKM2 has four domains, we generated four different domain-deletion PKM2 mutants (Fig. 6P) and transfected them into HL-1 cardiomyocytes. Co-IP assays showed that PKM2ΔA, PKM2ΔC, and PKM2ΔN, but not PKM2ΔB, were able to pull-down PHB2 (Fig. 6Q).
PKM2 induces PHB2 phosphorylation
After we revealed an interaction between PKM2 and PHB2, we asked whether conformational changes occur in PHB2 for its stabilization after PKM2 binding. To address this question, we analyzed the regions of PHB2 that are required for its interaction with PKM2. As PHB2 has four major domains, we generated four different domain deletion PHB2 mutants (Fig. 7A) and transfected them into HL-1 cardiomyocytes. Co-IP assays showed that PHB2ΔN, PHB2ΔCC, and PHB2ΔC, but not PHB2ΔPHB, were able to pull-down PKM2 (Fig. 7B). This suggests that the PHB domain of PHB2 determines its binding to PKM2.
PKM2 induces PHB2 phosphorylation. PKM2 transgenic (PKM2Tg) mice and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. In vivo measurements were performed after 24 h later, and mice administered an equal volume of phosphate buffer saline served as controls. Immortalized mouse cardiac muscle HL-1 cells were treated with 10 µg/mL of LPS for 24 h to simulate SC in vitro. Cells treated with an equal volume of phosphate buffer saline were used as controls. Before LPS treatment, cardiomyocytes were transduced with Adenovirus encoding PKM2 (Ad-PKM2). β-gal-overexpressing (Ad-β-gal) cells were used as controls. (A) Mapping of PHB2 regions and deletion mutants. (B) Representative immunoblot from immunoprecipitation analysis of HL-1 cells transfected with PHB2 deletion mutants. (C-F) Co-IP analysis of the interaction between dimeric and tetrameric PKM2 forms and PHB2 in HL-1 cells. (G-I) Western blot analysis of p-PHB2S176 and p-PHB2Ser91 expression in cardiac tissues. (J, K) Representative results of an in vitro kinase assay evaluating binding of phospho-PHB2 variants to PKM2 in the presence of LPS or compound 3k, a PKM2 inhibitor. (L, M) Pulse-chase analysis of PHB2 protein half-life in HL-1 cells transfected with PHB2S91D and PHB2S91A mutant constructs. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
PKM2 has two conformations, a tetrameric one with high protein kinase (PK) activity that catalyzes the production of pyruvate from phosphoenolpyruvate (PEP), and a dimeric one, which has lower PK activity and also exerts non-glycolytic enzymatic functions. Interestingly, co-IP results showed that PHB2 mainly interacted with dimeric, rather than tetrameric, PKM2 (Fig. 7C and F). Thus, LPS seems to interrupt the cooperation between PHB2 and dimeric PKM2. These results hinted that the kinase function of PKM2 is involved in the regulation of the PHB domain of PHB2. PKM2 has been reported to directly induce post-transcriptional phosphorylation of several proteins, such as ERK2T202 [37], β-cateninY333 [38], histone H3T11 [39], myosin light chain 2Y118 [40], and protein kinase B substrate l (AKT1S1)S202 [41]. On the other hand, PHB2 possesses three phosphorylation sites (S91, S176, and S243) which have been reported to enhance the anti-oxidative and anti-apoptotic properties of PHB2 through an undefined mechanism [42,43,44]. Since the PHB domain of PHB2 encompasses amino acids 68–185, we asked whether PKM2/PHB2 interaction results in PHB2 phosphorylation within this sequence fragment. Western blotting showed abundant expression of p-PHB2S91 and little p-PHB2S176 in heart tissues under baseline conditions. Neither LPS nor PKM2 overexpression had an impact on p-PHB2S176 expression (Fig. 7G and I). In contrast, after exposure to LPS the expression of p-PHB2S91 was downregulated in WT mice but remained at near normal levels in PKM2Tg mice, compared with PBS treated mice (Fig. 7G and I). An in vitro kinase assay further showed that PKM2 induced PHB2 phosphorylation at S91, and this alteration was significantly inhibited by compound 3k, a PKM2 inhibitor (Fig. 7J and K).
To evaluate whether PKM2-mediated PHB2 phosphorylation at S91 is necessary for PHB2 stabilization, phosphorylation-defective (PHB2S91A) and phosphorylation-mimetic (PHB2S91D) PHB2 mutants were transfected into HL-1 cells. After transfection with PHB2S91D, LPS-mediated PHB2 degradation was delayed in control cells (Fig. 7L and M). In turn, in cells transfected with a PHB2S91A mutant, PKM2 overexpression failed to maintain PHB2 stability (Fig. 7L and M). Taken together, our results showed that PKM2 binds to PHB2 and phosphorylates it at S91, leading to increased PHB2 stability.
PHB2 dephosphorylation abolishes PKM2-mediated mitochondrial protection
To evaluate whether PHB2 phosphorylation underlies PKM2-related MQC activation during SC, further assays were conducted in HL-1 cells transfected with PHB2S91A and PHB2S91D mutants. Immunofluorescent assessment of mitochondrial morphology showed that LPS-induced mitochondrial fragmentation was attenuated by overexpression of either PHB2S91D or PKM2 (Fig. 8A and C). However, in HL-1 cells transfected with PHB2S91A, PKM2 overexpression failed to maintain mitochondrial length and reduce the number of cardiomyocytes with plenty of fragmented mitochondria (Fig. 8A and C). Further, western blot analysis showed that LPS-mediated disruption of mitochondrial fission/fusion and mitophagy could be normalized by overexpression of PHB2S91D or PKM2 (Fig. 8D and J). In contrast, in HL-1 cells transfected with PHB2S91A, PKM2 overexpression failed to sustain mitochondrial fission/fusion and mitophagy (Fig. 8D and J). Similarly, mt-Keima assays confirmed that either PHB2S91D or PKM2 overexpression was able to sustain mitophagy in the presence of LPS, while PKM2-induced mitophagy was nullified by PHB2S91A transfection (Fig. 8K and L). Furthermore, mitochondrial biogenesis was stimulated by PHB2S91D or PKM2 overexpression in LPS-treated cells, while the effect of PKM2 overexpression was abrogated by PHB2S91A transfection (Fig. 8M and N). Lastly, transcriptions of mtUPR-related genes were slightly increased by LPS and partially restored toward baseline levels upon overexpression of either PHB2S91D or PKM2 (Fig. 8O-P). However, in cells transfected with the PHB2S91A mutant, the inhibitory effect of PKM2 on mtUPR was inhibited (Fig. 8O and P). These data indicate that PKM2-mediated mitochondrial protection occurs through PKM2-dependent PHB2 phosphorylation.
PHB2 dephosphorylation abolishes PKM2-mediated mitochondrial protection. HL-1 cardiomyocytes were transfected with PKM2 overexpression adenovirus (Ad-PKM2), a phosphorylation-defective PHB2S91A mutant, or a phosphorylation-mimetic PHB2 S91D mutant before LPS treatment. Adenovirus loaded β-gal (Ad-β-gal) cells were used as controls. (A-C) Immunofluorescence analysis of mitochondrial morphology in HL-1 cells. Representative images of mitochondria immunofluorescence (A), average mitochondrial length (B), and proportion of cardiomyocytes with fragmented mitochondria (C) are shown. (D-J) Western blot analysis of Drp1, Fis1, Mfn2, Opa1, Parkin, and Atg5 in HL-1 cells. (K, L) Mitophagy analysis results (mt-Keima assay). (M-P). Transcriptional analysis of Tfam, Nrf2, mtHsp70, and Atf6 expression by qPCR. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
PKM2-mediated cardiomyocyte protection against septic insult requires PHB2 phosphorylation
To illustrate the necessary role of PHB2 phosphorylation in supporting PKM2-induced cardioprotection, HL-1 cardiomyocyte viability and function were determined after transfection with PHB2S91D or PHB2S91A mutants. LPS exposure promoted the release of cardiac injury markers, e.g. TnI, CK-MB, and LDH, into the culture medium in control cells, and this trend was decreased by either PKM2 overexpression or introduction of the phospho-mimetic PHB2S91D mutant (Fig. 9A and C). In line with the findings described above, PKM2 overexpression failed to prevent the upregulation of cardiac damage markers in HL-1 cells co-transfected with the phospho-defective PHB2S91A mutant after LPS challenge (Fig. 9A and C). In turn, both cell viability and survival assessed by CCK-8 assay (Fig. 9D) and TUNEL staining (Fig. 9E and F), respectively, were improved by either PKM2 overexpression or PHB2S91D mutant transfection. In contrast, PKM2 overexpression-mediated cardiomyocyte survival was decreased in cells co-expressing PHB2S91A (Fig. 9D and F). Moreover, LPS-stimulated transcription of pro-inflammatory factors was significantly suppressed by either PKM2 or PHB2S91D overexpression, while the anti-inflammatory action of PKM2 was impaired upon transfection of PHB2S91A (Fig. 9G-H). Finally, either PKM2 or PHB2S91D, but not PHB2S91D transfection, largely preserved myosin filament expression and organization in cardiomyocytes exposed to LPS (Fig. 9I-J). The above evidence thus confirms that PKM2-mediated cardioprotection during septic conditions requires PHB2 phosphorylation.
PKM2-mediated cardiomyocyte protection against septic insult requires PHB2 phosphorylation. HL-1 cardiomyocytes were transfected with PKM2 overexpression Adenovirus (Ad-PKM2), a phosphorylation-defective PHB2S91A mutant, or a phosphorylation-mimetic PHB2 S91D mutant before LPS treatment. Adenovirus loaded β-gal (Ad-β-gal) cells were used as controls. (A, C) ELISA-based analysis of TnI, CK-MB, and LDH levels in culture media of HL-1 cells transfected with phosphorylation-defective (PHB2S91A) and phosphorylation-mimetic (PHB2S91D) mutant constructs. (D) Cell viability analysis via CCK-8 assay in vitro. (E, F) Apoptosis analysis by TUNEL staining in cultured HL-1 cells. (G, H) Transcriptional analysis of Tnfα and Mcp1 expression. (I, J) Representative images of myosin immunofluorescence. Myosin expression levels were normalized to those of the control group. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
Knockin mice expressing phospho-mimetic Phb2 S91D are less vulnerable to SC
To investigate whether PKM2-mediated PHB2 phosphorylation confers myocardial protection against SC in vivo, transgenic knockin mice carrying the Phb2S91D variant were generated. Total-PHB2 was used as the loading control to analyze the changes in p-PHB2S91. In comparison to WT mice, heart tissues from homozygous Phb2S91D/D mice showed elevated levels of p-PHB2S91 (Fig. 10A-B). Since PHB2 phosphorylation prevents PHB2 degradation, we further examined the alterations in total-PHB2 expression in WT, heterozygous Phb2S91D/+, and homozygous Phb2S91D/D mice. After normalizing to α-tubulin, the levels of total-PHB2 were significantly reduced in LPS-treated WT mice (Fig. 10C-D). However, these changes were partially prevented in Phb2S91D/+ mice and fully restored in Phb2S91D/D. Furthermore, the mtDNA/nDNA ratio was employed to assess potential alterations in mitochondrial content or mitochondrial copy numbers in transgenic knockin mice harboring the Phb2S91D variant. As depicted in Fig. 10E, the mtDNA/nDNA ratio exhibited no significant changes among WT, heterozygous Phb2S91D/+, and homozygous Phb2S91D/D mice.
Knockin mice expressing phospho-mimetic Phb2S91 are less vulnerable to SC. Heterozygous Phb2S91D/+ mice, homozygous Phb2S91D/D mice, and wild-type (WT) mice aged 8–10 weeks were injected intraperitoneally with 10 mg/kg LPS to induce SC. Mice administered an equal volume of phosphate buffer saline served as controls. (A, B) Western blot analysis of cardiac p-PHB2S91 in WT, heterozygous Phb2S91D/+, and homozygous Phb2S91D/D mice treated with PBS or LPS. Total-PHB2 was used as the loading control. (C, D) Western blot analysis of total-PHB2 in WT, heterozygous Phb2S91D/+, and homozygous Phb2S91D/D mice treated with PBS or LPS. α-tubulin was used as the loading control. (E) The CO1 gene of mtDNA and the NDUFV1 gene of nDNA were amplified using qPCR to assess the relative ratio of mtDNA/nDNA in heart tissues obtained from WT, heterozygous Phb2S91D/+, and homozygous Phb2S91D/D mice under normal physiological conditions. (F-L) Echocardiographic evaluation. LVDd, left ventricular diastolic dimension; LVDs, left ventricular systolic dimension; IVS, interventricular septum thickness; E/A, ratio of early to late transmitral flow velocities; FS, ratio of left ventricular fractional shortening. (M-O) ELISA-based analysis of serum TnI, CK-MB, and LDH concentrations. (P-R) Transcriptional analysis of cardiac Mmp9, Mcp1, and Tnfα expression by qPCR. (S) Representative images of TnT and Gr-1 immunohistochemistry in cardiac sections. (T) Quantification of Gr-1-positive neutrophils in mouse heart tissues. (U) ELISA-based measurement of caspase-3 activity in heart tissues. Values are presented as mean ± SEM. For in vivo data, n = 6 mice per group. For in vitro data, n = 4 independent experiments. #p < 0.05, and ##p < 0.01
Heart function, assessed by echocardiography, was slightly improved in Phb2S91D/+ mice (Fig. 10F and L), and largely normalized in Phb2S91D/D mice upon LPS-induced SC. Similarly, LPS-stimulated TnI, CK-MB, and LDH were partially reduced in Phb2S91D/+ mice and significantly inhibited in Phb2S91D/D mice (Fig. 10M and O). Along with these changes, LPS-induced transcription of Mmp9, Mcp1, and Tnfα was downregulated in Phb2S91D/+ mice, and further repressed in Phb2S91D/D mice (Fig. 10P and R). In accordance with these alterations, myocardial accumulation of Gr-1 positive neutrophils was relived in Phb2S91D/+ mice and completely prevented in Phb2S91D/D mice (Fig. 10S and T). Lastly, LPS-induced cardiomyocyte death, as assessed by caspase-3 activity, was partly attenuated in Phb2S91D/+ mice and remarkably inhibited in Phb2S91D/D mice (Fig. 10U). Taken together, our results elucidated that PHB2 phosphorylation mediates myocardial protection against LPS-induced SC.
Discussion
The present study focused on the role of PKM2 in cardiomyocyte homeostasis and MQC regulation in the setting of SC. Our main findings showed that: (1) LPS reduced the levels of myocardial PKM2 mainly through suppressed expression of dimeric PKM2; (2) PKM2 overexpression in mice correlated with reduced myocardial inflammation and cardiomyocyte death after SC induction; (3) overexpression of PKM2 improved heart performance through repressing LPS-triggered mitochondrial damage; (4) PKM2-mediated mitochondrial protection in the presence of LPS correlates with normalized MQC, as evidenced by balanced mitochondrial fission/fusion events, activated mitophagy, increased mitochondrial biogenesis, and mitigated mtUPR; (5) upon LPS stress, PKM2 stimulated MQC through sustaining PHB2 expression; (6) PKM2 interacts directly with PHB2 and induces PHB2 phosphorylation at Ser91, resulting in increased PHB2 stability; (7) the protective actions of PKM2 on cardiomyocyte homeostasis and mitochondrial integrity are nullified upon expression of a phosphorylation-defective PHB2S91A mutant construct. This study thus identified PHB2 dephosphorylation, leading to enhanced PHB2 protein degradation, as an initial signal determining MQC disruption during development of SC. Besides, we also showed that stabilized PKM2 expression is a prerequisite to sustain PHB2 phosphorylation after septic insult. These findings indicate that PKM2 downregulation, PKM2-PHB2 disassociation, PHB2 dephosphorylation, and MQC dysfunction are important determinators of sepsis-triggered myocardial injury.
Increasing evidence supports the beneficial effects offered by PKM2 in cardiovascular disorders. During cardiac development, loss of PKM2 in cardiomyocytes impairs cell cycling and reduces cardiomyocyte numbers and myocardium size [30]. By comparison, overexpression of PKM2 in heart tissues via cardiomyocyte-specific Pkm2 modified RNA protected the myocardium against acute or chronic myocardial infarction through enhancing heart function, inhibiting cell death, and repressing oxidative stress [30]. Mechanistically, PKM2-mediated cardioprotection seems to correlate with activation of anabolic pathways and β-catenin signaling, suggesting the dimeric PKM2, rather than tetrameric PKM2, is essential for maintaining heart performance [30]. Apart from its actions on cardiomyocytes, pharmacological activation of PKM2 in endothelial cells promotes proliferation and induces angiogenic differentiation [26]. Instead of mediating metabolic adaption, this beneficial effect has been reported to be associated with increased mitochondrial fusion/fission ratio, an effect accompanied by stabilized mitochondrial potential, decreased mitochondrial ROS generation, and angiogenesis-related gene expression [26]. Importantly, pulmonary hypertension [45], angiotensin II-triggered cardiac remodeling [46], and diabetic cardiomyopathy [47] were all shown to be relieved by PKM2 activation, either by genetic modification or pharmacological manipulation, through multiple mechanisms such as inhibition of TGF-β/Smad2/3 and Jak2/Stat3 pathways, oxidative stress, and NLRP3 inflammasome. These results highlight an unforeseen non-metabolic regulatory effect of PKM2 in cardioprotection. In accordance with the above results, our data also showed that SC is featured by a drop in cardiac levels of dimeric PKM2 and that re-introduction of PKM2 improves heart function through sustaining MQC. However, strategies aimed at modulating PKM2 expression require cautiousness, as the impact of its tetrameric and dimeric forms on heart performance would be different. Tetrameric PKM2 would increase glycolysis, a process less efficient than mitochondrial respiration in terms of ATP yield per glucose, despite demanding lower oxygen consumption. Of note, elevated PKM2-mediated glycolysis may result in lactic acidosis, a life-threatening medical condition during sepsis. Therefore, differential induction of dimeric PKM2 expression may be a safer approach to elicit cardioprotection.
A variety of regulatory mechanisms have been proposed to explain PHB2-mediated MQC control, including effects on total protein expression, post-transcriptional phosphorylation, and intracellular compartmentation. A reduction in total PHB2 expression in cardiac-specific PHB2 knockout mice impairs myocardial fatty acid oxidation (FAO) and induces heart failure through suppressing carnitine palmitoyltransferase 1b (CPT1b), a rate-limiting enzyme of cardiac mitochondrial FAO [48]. On the contrary, upregulation of PHB2 expression inhibits cardiomyocyte senescence and therefore delays heart aging through increased mitophagic flux [49]. In addition to altered PHB2 protein expression, phosphorylation of PHB2 at Ser91 is required for myocytic differentiation through MEF2-dependent transcription [44]. Meanwhile, experiments in leukemia cells showed that PHB2 phosphorylation at either S176 or S91 exerts anti-apoptotic actions by attenuating mitochondrial dysfunction [43]. In the nucleus, PHB2 regulates the expression of transcription factors involving in cell cycle progression [42], and increased nuclear PHB2 expression has been regarded as an early event for mitochondrial cristae disarrangement and disruption of inner mitochondrial membrane integrity [50]. Consistent with these findings, we found that experimental SC induces PHB2 downregulation and dephosphorylation in mouse cardiomyocytes. Importantly, our data further suggest that PHB2 dephosphorylation constitutes an endogenous mechanism inducing PHB2 degradation and downregulation. In fact, it is recognized that phosphorylation impacts protein degradation in a site-specific manner, influenced by surrounding amino acid composition, local structure, and phosphoprotein function [51]. In this regard, several proteins, including WRKY [52], p300 [53], c-Myb [54], and RhoA [55], may be degraded through post-transcriptional phosphorylation. Hence, our findings may serve to establish a novel cause-effect relationship between PHB2 phosphorylation and its total expression.
Limitation
Our present study has a few limitations that deserve mention. Firstly, although we observed that LPS had complex effect on PHB2 expression, additional experiments are required to figure out the mechanism by which LPS induces PHB2 degradation. Secondly, the evaluation of mitochondrial function primarily relied on in vivo experiments, with only a limited number of in vitro experiments analyzing mitochondrial respiration. Consequently, a more comprehensive assessment of mitochondrial function using additional in vitro techniques would strengthen our findings. Besides, the interactive effects between PKM2 and PHB2 were primarily examined in vitro. To better elucidate the underlying interplay mechanism of PKM2-mediated PHB2 phosphorylation, it is essential to conduct animal studies using genetically modified mice. Our present study has a few limitations that deserve mention. Firstly, although we observed that LPS had complex effect on PHB2 expression, additional experiments are required to figure out the mechanism by which LPS induces PHB2 degradation. Secondly, the evaluation of mitochondrial function primarily relied on in vivo experiments, with only a limited number of in vitro experiments analyzing mitochondrial respiration. Consequently, a more comprehensive assessment of mitochondrial function using additional in vitro techniques would strengthen our findings. Besides, the interactive effects between PKM2 and PHB2 were primarily examined in vitro. To better elucidate the underlying interplay mechanism of PKM2-mediated PHB2 phosphorylation, it is essential to conduct animal studies using genetically modified mice. Lastly, it appears that an additional regulatory mechanism is involved in the phosphorylation of PHB2 in Phb2D/D knock-in mice during LPS exposure, as compared to LPS-treated WT mice and a phosphor-mimetic in vivo mutant seems to be attenuated by LPS due to undefined mechanism. We acknowledge these limitations and believe that addressing them in future studies will provide a more comprehensive understanding of the mechanisms involved.
Conclusion
In summary, our study highlights a novel mechanism by which PKM2 downregulation impairs MQC in cardiomyocytes during SC. Specifically, our results show that PKM2-mediated phosphorylation of PHB2 at Ser91 is a key upstream event that serves to normalize mitochondrial homeostasis under septic challenge. Our findings may lay the foundation for research and development of new drugs aimed at preventing MQC dysfunction in SC by targeting the PKM2/PHB2 axis.
Materials and methods
Septic cardiopathy mouse model
C57BL/6J wild-type (WT) male mice (8–12 weeks old) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). PKM2 transgenic (PKM2Tg) mice were generated by Cyagen Biosciences (Santa Clara, CA, USA) according to a previous study [31] with a minor modification. In brief, mouse full-length PKM2 cDNA was amplified, ligated with a pGEMT-Easy vector, and transformed in NEB 5-α competent E. coli. Purified transgene constructs were microinjected into C57BL/6J egg pronuclei to generate PKM2Tg mice. PHB2S91D-knockin mice on a C57BL/6 background were also generated by Cyagen Biosciences. All mice were born and maintained under pathogen-free conditions. Male mice were used to induce SC. LPS (E. coli 0111: B4, #2630, Sigma-Aldrich, USA) was dissolved in phosphate buffered saline at 10 mg/kg and administered to mice intraperitoneally. Systolic blood pressure was determined by a noninvasive tail-cuff instrument (XBP 1000; Kent Scientific, Torrington, CT, USA). After 24 h, mice were sacrificed and serum was collected to analyze white blood cells (WBC), neutrophils, and monocyte numbers using an automatic blood analyzer (Sysmex XT-2000i; Sysmex Corp., Shanghai, China).
Echocardiography
Echocardiograms were obtained with a Vevo 2100 Ultrasound System (FUJIFILM VisualSonics Inc., Toronto, Ontario, Canada) equipped with a high-frequency (30 MHz) linear array transducer. Mice were lightly anesthetized with isoflurane (3% for induction and 1.0-1.5% for maintenance) mixed with 1 L/min O2 via a facemask [56]. Hair was removed from the chest using a chemical hair remover, and the animals were placed in the left lateral decubitus position. During measurements, body temperature was carefully monitored with a rectal temperature probe and maintained close to 37 °C with the use of a heating pad. Ultrasound gel was applied to the chest and two-dimensional, M-mode, and spectral Doppler images were acquired from modified parasternal long-axis and short-axis views. All images were digitally stored for off-line analysis using Vevo LAB software (version 1.7.1, VisualSonics). Systolic and diastolic anatomic measurements were obtained from M-mode images at the mid-papillary level. LV mass was estimated by the area-length method. Analysis was performed by an investigator who was blind to treatment allocation and unaware of data from other modalities [57].
Histological analysis and immunohistochemistry
Cardiac mouse tissue was fixed, embedded in paraffin, cut into 4 μm-thick sections, and stained with hematoxylin and eosin (HE). For ICAM1 and VCAM1 immunohistochemistry, 5 μm-thick frozen heart sections were prepared on a cryostat, fixed with methanol/acetone, treated with 3% H2O2 to quench endogenous peroxidase activity, blocked with 2% bovine serum albumin (BSA), and incubated with anti-VCAM1 (1:500; #ab215380; Abcam, Cambridge, UK) and anti-ICAM1 (1:500; #ab171123; Abcam) antibodies overnight at 4 °C. After extensive washing, sections were incubated with suitable biotinylated secondary antibodies (ABC Kit, MX-200-4; Agilent Technologies, Santa Clara, CA, USA) [58]. Positive staining was visualized using DAB substrate (Solarbio, Beijing, China).
Immunofluorescence
Heart sections were prepared according to a standard procedure and then incubated with blocking buffer containing 10% (w/v) BSA. Afterward, the sections were incubated overnight at 4 °C with primary antibodies and subsequently with a fluorophore-conjugated secondary antibody (Alexa Fluor® 568 goat anti-rabbit IgG (H + L); A11036; Invitrogen, Waltham, MA, USA) for 1 h. Nuclei were then stained with DAPI and images captured with a fluorescence microscope (BX51, Olympus, Japan). The primary antibodies were listed in the Supplementary Table 1.
Electron microscopy
Transmission electron microscopy was performed according to a previous report [59]. In brief, samples were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 for 1 h, and then washed in 100 mM phosphate buffer without CaCl2. All samples were post-fixed in 1% osmium tetroxide for 1 h and stained in 2% uranyl acetate in maleate buffer pH 5.2 for an additional hour. The samples were rinsed, dehydrated in an ethanol series, embedded in resin (EMbed 812, EMS, Hatfield, PA, USA), and baked overnight at 60 °C. Hardened blocks were cut using a Leica Ultra Cut UCT microtome. Ultrathin Sect. (60 nm) were collected on carbon-coated grids and contrast-stained using 2% uranyl acetate and lead citrate. Embedded samples were analyzed by a JEM-1400 Plus electron microscope at 80 KV (Jeol Ltd., Tokyo, Japan).
Primary cardiomyocyte culture and treatment
Mouse hearts were removed and perfused using a Langendorff system for ∼ 3 min with Ca2+-free bicarbonate-based buffer. Enzymatic digestion was initiated by adding collagenase type B/D to the perfusion solution. After approximately 7 min, the left ventricle was removed and discarded. Male mice cardiomyocytes from the left ventricular wall were isolated as reported previously by us [60] and plated at 1 ∼ 2 × 104 cells/cm2 in mouse laminin pre-coated culture dishes. After 1 h culturing in a 5% CO2 incubator at 37 °C, cardiomyocytes were incubated with Fura 2-AM (Invitrogen) for 10 min and then subjected to field stimulation at 2 Hz with platinum electrodes. Contraction and relaxation measures were determined as previously described [59]. Additional experiments were performed using the cardiac mouse HL-1 cell line (American Type Culture Collection, Manassas, VA, USA). To induce inflammation-mediated damage, HL-1 cells were treated with 10 µg/ml LPS for 24 h [35].
RNA extraction, reverse transcription, and qRT-PCR
Total RNA extraction was performed on heart tissues and HL-1 cells using TRIzol® reagent (Ambion® by Life Technologies, Austin, TX, USA) [14]. Purified RNA samples were reverse transcribed for cDNA synthesis using the GeneAmp™ RNA PCR Core Kit (Applied Biosystems, Waltham, MA, USA). qRT-PCR amplification of target genes and GAPDH (internal control) from heart tissues was performed using the TaqMan® Gene Expression Assay (Applied Biosystems). Amplification of target genes and GAPDH from HL-1 cells was performed using SYBR™ Green qPCR Master Mix (Applied Biosystems). The primers used are listed in Supplementary Table 2. An StepOnePlus™ Fast Real-Time PCR System (Applied Biosystems) was used for all qRT-PCR assays. The Cytochrome c oxidase subunit I (CO1) gene of the mtDNA was amplified by qPCR to determine the mtDNA copy. The transcript level of mtDNA was reflected by NADH dehydrogenase subunit 1 (ND1). The CO1 gene, representing the cytochrome c oxidase subunit I in mtDNA, and the NDUFV1 gene in nDNA were amplified using qPCR. The amplification process began with an initial step at 94 °C for 10 min, followed by 40 cycles consisting of 94 °C for 10 s, 60 °C for 30 s, and 94 °C for 10 s. All reactions were performed in duplicate. Subsequently, the amplification curves were analyzed using SDS 1.9.1 software (Applied Biosystems). Based on a previous study [61], these curves were utilized to determine the relative mtDNA: nDNA ratio in each sample.
Protein extraction and western blotting
Whole-cell protein lysates were extracted from heart tissues or HL-1 cells using RIPA lysis buffer (150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris, pH 8.0) supplemented with Complete Protease and PhosSTOP phosphatase inhibitors (both from Roche Diagnostics, Basel, Switzerland) [62]. To prepare mitochondrial protein lysates, mitochondrial fractions from human HL-1 cells were isolated using the Mitochondria Isolation Kit for Mammalian Cells (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Equal volumes of protein lysates were separated by SDS-PAGE and transferred onto PVDF membranes (GE Healthcare, Little Chalfont, UK). The antibodies used for western blotting are listed in Supplementary Table 1.
Seahorse assay
The Seahorse XFe96 extracellular flux analyzer (Agilent) was used to assess respiration and mitochondrial function. Cells were seeded on Matrigel-coated assay plates 7 days before measurement at a density of 20,000 cells per XFe96 well. Cells were washed twice in Agilent Seahorse XFe DMEM Basal Medium (Agilent Technologies, cat. no. 103575-100) supplemented with 2 mM glutamine, 10 mM glucose and 1 mM sodium pyruvate 1 h before the assay and for the duration of the measurement. For the standard profiling (Mito Stress test), oligomycin was injected at 1.5 µM, FCCP at 0.5 µM and rotenone/antimycin A were added at 0.5 µM. The oxygen consumption rate (OCR) was normalized to the number of nuclei quantified by DAPI staining. OCR and ATP production rates were obtained using the Seahorse Wave controller Software 2.6.1 (Agilent) as our previously described [63].
Co-immunoprecipitation
Co-immunoprecipitation (co-IP) was performed as described previously [20]. Briefly, HL-1 cells were harvested and then solubilized with IP buffer (150 mM NaCl, 10% glycerol, 20 mM Tris–HCl pH 7.4, 2 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100, and complete protease inhibitor) for 1.5 h at 4 °C. The insoluble material was removed by centrifugation at 12,000 × g for 10 min, and the supernatant was collected. One-tenth of the supernatant was pipetted out and used as ‘Input’. The remaining supernatant fraction was subsequently incubated with anti-Flag M2 affinity gel (Sigma-Aldrich) at 4 °C overnight. The beads were washed six times with lysis buffer, and the proteins (IP products) were eluted by boiling the beads in SDS sample buffer. Finally, the samples were analyzed by western blotting [64].
In vitro kinase assay
In vitro kinase assays were conducted at 27 °C for 90 min in 20 mM HEPES pH 7.0, 1 mM TCEP, 10 mM MgCl2, 0.1 mM Na3VO4, and 0.6 µM BSA, with 1.6 µM recombinant mouse PHB2 protein (#MBS1037998; MyBioSource, Inc., San Diego, CA, USA), 12.5 µM ATP, 2 nM recombinant mouse PKM2 protein (#ab95474, Abcam), and 20 nM ADP-heptose or 20 nM DF-006 A, according to our previous reports [65].
Analysis of myocardial inflammation and myocardial injury markers
Quantitative measurements of myocardial inflammation and myocardial injury biomarkers in cell culture medium or mouse serum were performed using mouse ELISA kits: Serum Creatine Kinase MB (CK-MB) (#E4607; BioVision, Waltham, MA, USA), Troponin I (TNI) (#MBS854284; MyBioSource, Inc.), and Lactate Dehydrogenase (LDH) (#MBS720560; MyBioSource, Inc.) [66]. Apoptosis induction was evaluated with a Caspase-3 Colorimetric Assay Kit (#K106; BioVision, Inc.) and a Caspase-9 Colorimetric Assay Kit (#K119; BioVision, Inc.), as per manufacturer’s instructions [67].
Mitochondrial membrane potential, mitochondrial ROS, and mitochondrial permeability transition pore (mPTP) detection
For mitochondrial membrane potential quantification, HL-1 cells were incubated in media containing 10 nM JC-1 (#E-CK-A301; Elabscience, Houston, TX, USA) at 37 °C for 10 min. Images were captured under a fluorescence microscope (BX51, Olympus, Japan). MitoSOX™ Red Mitochondrial Superoxide Indicator (#M36008; Thermo Fisher Scientific) was used to evaluate the production of mitochondrial ROS (mROS) in cells under LPS treatment. In brief, cells were incubated with 5 nM MitoSOX™ Red at 37 °C for 10 min and then observed using a fluorescence microscope (BX51, Olympus, Japan). mPTP detection was performed using an Image-IT™ LIVE Mitochondrial Transition Pore Assay Kit (#I35103, Thermo Fisher Scientific) as per manufacturer’s instructions [68].
Adenovirus vector construction
A PKM2 overexpression plasmid was cloned into pcDNA3.1 (Invitrogen) vector for adenovirus production. PHB2 sequences were translated from HL-1 cell cDNA. PHB2 has four domains, including an N-terminal mitochondrial targeting domain (N, 1–50 aa), a PHB domain (PHB, 68–185 aa), a coiled coil domain (CC, 190–264 aa), and a C-terminal region (C, 265–299 aa). Full length PHB2, PHB2 without N-terminal domain (PHB2ΔN), PHB2 without C-terminal domain (PHB2ΔC), PHB2 without CC domain (PHB2ΔCC), and PHB2 without PHB domain (PHB2ΔPHB) were generated by PCR and ligated into pHAGE-HA vectors for adenovirus production. PKM2 contains an N-terminal domain (N, 1–43 aa), an A domain (44–116 aa and 219–389 aa), a B domain (117–218 aa), and a C region (390–531 aa). Full length PKM2, PKM2 without N domain (PKM2ΔN), PKM2 without A domain (PKM2ΔA), PKM2 without C domain (PKM2ΔC), and PKM2 without N domain (PKM2ΔN) were generated by PCR and ligated into pHAGE-Myc vectors for adenovirus production. The constitutively active (PHB2S91D) and inactive (PHB2S91A) PHB2 mutant plasmids were obtained to construct lenti-PHB2 viruses as previously described by us [69]. The adenovirus was packaged in 293T cells using Opti-MEM reduced serum medium, along with helper plasmids and PEI reagent. adenovirus supernatant was collected 48 h later and used to infect HL-1 cells after addition of polybrene. Positive clones were then selected by puromycin [69].
Pulse-chase analysis
Protein stability was determined through pulse-chase analysis as described by us previously [59]. In brief, HL-1 cells were first radiolabeled with [35 S]-Met (100 mCi) under complete medium for the indicated times at 37 °C in the absence or presence of LPS. After washing twice with PBS, cells were cultured under complete medium for the indicated duration. Then, cell samples were lysed and immunoprecipitated with protein G Plus beads coated with the indicated antibodies, followed by western blot analysis [70].
Cell viability
Cell viability was determined using a Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions [71]. HCC cells were seeded in 96-well plates (2 × 103 cells per well) in a final volume of 100 µl. At the indicated time point, 10 µl of CCK-8 reagent was added into each well for 1 h at 37 °C, and absorbance was measured at 450 nm using a multimode plate reader (BioTek, USA).
Statistical analysis
Data were analyzed using GraphPad Prism 5 software and are shown as mean values and SEM. Differences between two groups were analyzed by unpaired t test if data were normally distributed. Otherwise, Mann-Whitney U test was used. One or two-way ANOVA was used for comparisons between more than two groups. Bonferroni post-hoc test was further performed to compare two specific groups. Two-tailed tests were performed in all statistical analyses, and p < 0.05 was considered significant.
Data availability
The data supporting the findings of this study are found within the article and the supplementary material. All relevant raw data will be made available from the corresponding author upon reasonable request.
References
Montero S, Abrams D, Ammirati E, Huang F, Donker DW, Hekimian G, García-García C, Bayes-Genis A, Combes A, Schmidt M (2022) Fulminant myocarditis in adults: a narrative review. J Geriatr Cardiol 19:137–151
Zhao YW, Yan KX, Sun MZ, Wang YH, Chen YD, Hu SY (2022) Inflammation-based different association between anatomical severity of coronary artery disease and lung cancer. J Geriatr Cardiol 19:575–582
Frustaci A, Russo MA, Chimenti C (2009) Randomized study on the efficacy of immunosuppressive therapy in patients with virus-negative inflammatory cardiomyopathy: the TIMIC study. Eur Heart J 30:1995–2002
Huang BT, Yang L, Yang BS, Huang FY, Xiao QF, Pu XB, Peng Y, Chen M (2022) Relationship of body fat and left ventricular hypertrophy with the risk of all-cause death in patients with coronary artery disease. J Geriatr Cardiol 19:218–226
Haileselassie B, Mukherjee R, Joshi AU, Napier BA, Massis LM, Ostberg NP, Queliconi BB, Monack D, Bernstein D, Mochly-Rosen D (2019) Drp1/Fis1 interaction mediates mitochondrial dysfunction in septic cardiomyopathy. J Mol Cell Cardiol 130:160–169
Jiang X, Cai S, Jin Y, Wu F, He J, Wu X, Tan Y, Wang Y (2021) Irisin Attenuates Oxidative Stress, Mitochondrial Dysfunction, and Apoptosis in the H9C2 Cellular Model of Septic Cardiomyopathy through Augmenting Fundc1-Dependent Mitophagy. Oxid Med Cell Longev 2021:2989974
Bonanad C, Fernández-Olmo R, García-Blas S, Alarcon JA, Díez-Villanueva P, Mansilla CR, García-Pardo H, Toledo P, Ayesta A, Pereira E, Carol A, Castro-Conde A, De Pablo-Zarzoso C, Martínez-Sellés M, Arrarte V, Campuzano R, Ariza-Solé A (2022) Cardiovascular prevention in elderly patients. J Geriatr Cardiol 19:377–392
Shang X, Lin K, Zhang Y, Li M, Xu J, Chen K, Zhu P, Yu R (2020) Mst1 deletion reduces septic cardiomyopathy via activating parkin-related mitophagy. J Cell Physiol 235:317–327
Luo FY, Bai YP, Bu HS (2022) Protein quality control systems in hypertrophic cardiomyopathy: pathogenesis and treatment potential. J Geriatr Cardiol 19:780–784
Chang X, Toan S, Li R, Zhou H (2022) Therapeutic strategies in ischemic cardiomyopathy: focus on mitochondrial quality surveillance. EBioMedicine 84:104260
Chang X, Lochner A, Wang HH, Wang S, Zhu H, Ren J, Zhou H (2021) Coronary microvascular injury in myocardial infarction: perception and knowledge for mitochondrial quality control. Theranostics 11:6766–6785
Wang J, Zhou H (2020) Mitochondrial quality control mechanisms as molecular targets in cardiac ischemia-reperfusion injury. Acta Pharm Sin B 10:1866–1879
Wang J, Toan S, Zhou H (2020) Mitochondrial quality control in cardiac microvascular ischemia-reperfusion injury: new insights into the mechanisms and therapeutic potentials. Pharmacol Res 156:104771
Zhu H, Tan Y, Du W, Li Y, Toan S, Mui D, Tian F, Zhou H (2021) Phosphoglycerate mutase 5 exacerbates cardiac ischemia-reperfusion injury through disrupting mitochondrial quality control. Redox Biol 38:101777
Tan Y, Mui D, Toan S, Zhu P, Li R, Zhou H (2020) SERCA overexpression improves mitochondrial Quality Control and attenuates Cardiac Microvascular Ischemia-Reperfusion Injury. Mol Ther Nucleic Acids 22:696–707
Yu LM, Dong X, Xue XD, Xu S, Zhang X, Xu YL, Wang ZS, Wang Y, Gao H, Liang YX, Yang Y, Wang HS (2021) Melatonin attenuates diabetic cardiomyopathy and reduces myocardial vulnerability to ischemia-reperfusion injury by improving mitochondrial quality control: role of SIRT6. J Pineal Res 70:e12698
Xiong Z, Xing C, Xu T, Yang Y, Liu G, Hu G, Cao H, Zhang C, Guo X, Yang F (2021) Vanadium induces oxidative stress and mitochondrial Quality Control Disorder in the heart of ducks. Front Vet Sci 8:756534
Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF (2014) CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5:3596
Signorile A, De Rasmo D, Cormio A, Musicco C, Rossi R, Fortarezza F, Palese LL, Loizzi V, Resta L, Scillitani G, Cicinelli E, Simonetti F, Ferretta A, Russo S, Tufaro A, Cormio G (2019) Human ovarian Cancer tissue exhibits increase of mitochondrial Biogenesis and Cristae Remodeling. Cancers (Basel) 11
Yan C, Gong L, Chen L, Xu M, Abou-Hamdan H, Tang M, Désaubry L, Song Z (2020) PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 16:419–434
Wei Y, Chiang WC, Sumpter R Jr., Mishra P, Levine B (2017) Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168:224–238e10
Liu L, Bai F, Song H, Xiao R, Wang Y, Yang H, Ren X, Li S, Gao L, Ma C, Yang X, Liang X (2022) Upregulation of TIPE1 in tubular epithelial cell aggravates diabetic nephropathy by disrupting PHB2 mediated mitophagy. Redox Biol 50:102260
Shi Y, Li Q, Sun F, Zhu C, Ma S, Qin D, Li Q, Li T (2020) Lamprey PHB2 maintains mitochondrial stability by tanslocation to the mitochondria under oxidative stress. Fish Shellfish Immunol 104:613–621
Zhang Z, Deng X, Liu Y, Liu Y, Sun L, Chen F (2019) PKM2, function and expression and regulation. Cell Biosci 9:52
Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452:230–233
Ren R, Guo J, Shi J, Tian Y, Li M, Kang H (2020) PKM2 regulates angiogenesis of VR-EPCs through modulating glycolysis, mitochondrial fission, and fusion. J Cell Physiol 235:6204–6217
Li D, Shen C, Liu L, Hu J, Qin J, Dai L, Gao L, Cheng M, Wang D, Bao R, Wang B (2022) PKM2 regulates cigarette smoke-induced airway inflammation and epithelial-to-mesenchymal transition via modulating PINK1/Parkin-mediated mitophagy. Toxicology 477:153251
Qi W, Keenan HA, Li Q, Ishikado A, Kannt A, Sadowski T, Yorek MA, Wu IH, Lockhart S, Coppey LJ, Pfenninger A, Liew CW, Qiang G, Burkart AM, Hastings S, Pober D, Cahill C, Niewczas MA, Israelsen WJ, Tinsley L, Stillman IE, Amenta PS, Feener EP, Vander Heiden MG, Stanton RC, King GL (2017) Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med 23:753–762
Ni L, Lin B, Hu L, Zhang R, Fu F, Shen M, Yang J, Shi D (2022) Pyruvate kinase M2 protects heart from pressure overload-Induced Heart failure by phosphorylating RAC1. J Am Heart Assoc 11:e024854
Magadum A, Singh N, Kurian AA, Munir I, Mehmood T, Brown K, Sharkar MTK, Chepurko E, Sassi Y, Oh JG, Lee P, Santos CXC, Gaziel-Sovran A, Zhang G, Cai CL, Kho C, Mayr M, Shah AM, Hajjar RJ, Zangi L (2020) Pkm2 regulates Cardiomyocyte Cell cycle and promotes Cardiac Regeneration. Circulation 141:1249–1265
Han J, Hyun J, Park J, Jung S, Oh Y, Kim Y, Ryu SH, Kim SH, Jeong EI, Jo DG, Park SH, Jung YK (2021) Aberrant role of pyruvate kinase M2 in the regulation of gamma-secretase and memory deficits in Alzheimer’s disease. Cell Rep 37:110102
Wang R, Xu Y, Fang Y, Wang C, Xue Y, Wang F, Cheng J, Ren H, Wang J, Guo W, Liu L, Zhang M (2022) Pathogenetic mechanisms of septic cardiomyopathy. J Cell Physiol 237:49–58
Zhou H, Ren J, Toan S, Mui D (2021) Role of mitochondrial quality surveillance in myocardial infarction: from bench to bedside. Ageing Res Rev 66:101250
Wang J, Toan S, Zhou H (2020) New insights into the role of mitochondria in cardiac microvascular ischemia/reperfusion injury. Angiogenesis 23:299–314
Wang Y, Jasper H, Toan S, Muid D, Chang X, Zhou H (2021) Mitophagy coordinates the mitochondrial unfolded protein response to attenuate inflammation-mediated myocardial injury. Redox Biol 45:102049
Signorile A, Sgaramella G, Bellomo F, De Rasmo D (2019) Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 8
Keller KE, Doctor ZM, Dwyer ZW, Lee YS (2014) SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell 53:700–709
Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z (2011) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480:118–122
Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150:685–696
Jiang Y, Wang Y, Wang T, Hawke DH, Zheng Y, Li X, Zhou Q, Majumder S, Bi E, Liu DX, Huang S, Lu Z (2014) PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat Commun 5:5566
He CL, Bian YY, Xue Y, Liu ZX, Zhou KQ, Yao CF, Lin Y, Zou HF, Luo FX, Qu YY, Zhao JY, Ye ML, Zhao SM, Xu W (2016) Pyruvate kinase M2 activates mTORC1 by phosphorylating AKT1S1. Sci Rep 6:21524
Bavelloni A, Piazzi M, Raffini M, Faenza I, Blalock WL (2015) Prohibitin 2: at a communications crossroads. IUBMB Life 67:239–254
Bavelloni A, Piazzi M, Faenza I, Raffini M, D’angelo A, Cattini L, Cocco L, Blalock WL (2014) Prohibitin 2 represents a novel nuclear AKT substrate during all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Faseb j 28:2009–2019
Sun L, Cao X, Liu B, Huang H, Wang X, Sui L, Yin W, Ma K (2011) CaMK IV phosphorylates prohibitin 2 and regulates prohibitin 2-mediated repression of MEF2 transcription. Cell Signal 23:1686–1690
Zhang H, Wang D, Li M, Plecitá-Hlavatá L, D’alessandro A, Tauber J, Riddle S, Kumar S, Flockton A, Mckeon BA, Frid MG, Reisz JA, Caruso P, El Kasmi KC, Ježek P, Morrell NW, Hu CJ, Stenmark KR (2017) Metabolic and proliferative state of vascular adventitial fibroblasts in pulmonary hypertension is regulated through a MicroRNA-124/PTBP1 (Polypyrimidine Tract Binding Protein 1)/Pyruvate kinase muscle Axis. Circulation 136:2468–2485
Zhang X, Zheng C, Gao Z, Wang L, Chen C, Zheng Y, Meng Y (2021) PKM2 promotes angiotensin-II-induced cardiac remodelling by activating TGF-β/Smad2/3 and Jak2/Stat3 pathways through oxidative stress. J Cell Mol Med 25:10711–10723
Li Q, Leng K, Liu Y, Sun H, Gao J, Ren Q, Zhou T, Dong J, Xia J (2020) The impact of hyperglycaemia on PKM2-mediated NLRP3 inflammasome/stress granule signalling in macrophages and its correlation with plaque vulnerability: an in vivo and in vitro study. Metabolism 107:154231
Wu D, Jian C, Peng Q, Hou T, Wu K, Shang B, Zhao M, Wang Y, Zheng W, Ma Q, Li CY, Cheng H, Wang X, Zhao L (2020) Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure. Cell Death Dis 11:181
Wang L, Tang XQ, Shi Y, Li HM, Meng ZY, Chen H, Li XH, Chen YC, Liu H, Hong Y, Xu HH, Liu L, Zhao L, Han WN, Liu X, Zhang Y (2022) Tetrahydroberberrubine retards heart aging in mice by promoting PHB2-mediated mitophagy. Acta Pharmacol Sin
Osman C, Merkwirth C, Langer T (2009) Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci 122:3823–3830
Wu C, Ba Q, Lu D, Li W, Salovska B, Hou P, Mueller T, Rosenberger G, Gao E, Di Y, Zhou H, Fornasiero EF, Liu Y (2021) Global and Site-Specific Effect of Phosphorylation on protein turnover. Dev Cell 56:111–124e6
Yamada Y, Sato F (2016) Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis. Sci Rep 6:31988
Wang SA, Hung CY, Chuang JY, Chang WC, Hsu TI, Hung JJ (2014) Phosphorylation of p300 increases its protein degradation to enhance the lung cancer progression. Biochim Biophys Acta 1843:1135–1149
Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, Matsumoto K, Ishii S (2004) Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev 18:816–829
Rolli-Derkinderen M, Sauzeau V, Boyer L, Lemichez E, Baron C, Henrion D, Loirand G, Pacaud P (2005) Phosphorylation of serine 188 protects RhoA from ubiquitin/proteasome-mediated degradation in vascular smooth muscle cells. Circ Res 96:1152–1160
Hang PZ, Li PF, Liu J, Li FF, Chen TT, Pan Y, Zhang MR, Yu HQ, Ji HY, Du ZM, Zhao J (2022) Small-molecule 7,8-dihydroxyflavone counteracts compensated and decompensated cardiac hypertrophy via AMPK activation. J Geriatr Cardiol 19:853–866
Yao Y, Zhu P, Xu N, Jiang L, Tang XF, Song Y, Zhao XY, Qiao SB, Yang YJ, Yuan JQ, Gao RL (2022) Effects of chronic obstructive pulmonary disease on long-term prognosis of patients with coronary heart disease post-percutaneous coronary intervention. J Geriatr Cardiol 19:428–434
Mene-Afejuku TO, Jeyashanmugaraja GP, Hoq M, Ola O, Shah AJ (2022) Determinants of mortality among seniors acutely readmitted for heart failure: racial disparities and clinical correlations. J Geriatr Cardiol 19:719–724
Zhou H, Toan S, Zhu P, Wang J, Ren J, Zhang Y (2020) DNA-PKcs promotes cardiac ischemia reperfusion injury through mitigating BI-1-governed mitochondrial homeostasis. Basic Res Cardiol 115:11
Zhou H, Zhu P, Wang J, Zhu H, Ren J, Chen Y (2018) Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2alpha-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ 25:1080–1093
Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH (2009) DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion 9:261–265
Zou RJ, Tao J, He J, Wang CJ, Tan ST, Xia Y, Chang X, Li RB, Wang G, Zhou H, Fan XP (2022) PGAM5-Mediated PHB2 Dephosphorylation Contributes to Diabetic Cardiomyopathy by Disrupting Mitochondrial Quality Surveillance. RESEARCH 2022
Zhou H, Du W, Li Y, Shi C, Hu N, Ma S, Wang W, Ren J (2018) Effects of melatonin on fatty liver disease: the role of NR4A1/DNA-PKcs/p53 pathway, mitochondrial fission, and mitophagy. J Pineal Res 64
Zou R, Shi W, Qiu J, Zhou N, Du N, Zhou H, Chen X, Ma L (2022) Empagliflozin attenuates cardiac microvascular ischemia/reperfusion injury through improving mitochondrial homeostasis. Cardiovasc Diabetol 21:106
Wang S, Zhu H, Li R, Mui D, Toan S, Chang X, Zhou H (2022) DNA-PKcs interacts with and phosphorylates Fis1 to induce mitochondrial fragmentation in tubular cells during acute kidney injury. Sci Signal 15:eabh1121
Tan Y, Zhang Y, He J, Wu F, Wu D, Shi N, Liu W, Li Z, Liu W, Zhou H, Chen W (2022) Dual specificity phosphatase 1 attenuates inflammation-induced cardiomyopathy by improving mitophagy and mitochondrial metabolism. Mol Metab 64:101567
Wang J, Zhu P, Toan S, Li R, Ren J, Zhou H (2020) Pum2-Mff axis fine-tunes mitochondrial quality control in acute ischemic kidney injury. Cell Biol Toxicol 36:365–378
Zhou H, Zhu P, Wang J, Toan S, Ren J (2019) DNA-PKcs promotes alcohol-related liver disease by activating Drp1-related mitochondrial fission and repressing FUNDC1-required mitophagy. Signal Transduct Target Ther 4:56
Wang J, Zhu P, Li R, Ren J, Zhang Y, Zhou H (2020) Bax inhibitor 1 preserves mitochondrial homeostasis in acute kidney injury through promoting mitochondrial retention of PHB2. Theranostics 10:384–397
Wang J, Zhu P, Li R, Ren J, Zhou H (2020) Fundc1-dependent mitophagy is obligatory to ischemic preconditioning-conferred renoprotection in ischemic AKI via suppression of Drp1-mediated mitochondrial fission. Redox Biol 30:101415
Zou R, Tao J, Qiu J, Lu H, Wu J, Zhu H, Li R, Mui D, Toan S, Chang X, Zhou H, Fan X (2022) DNA-PKcs promotes sepsis-induced multiple organ failure by triggering mitochondrial dysfunction. J Adv Res 41:39–48
Acknowledgements
None.
Funding
This study was supported by grants from Beijing Natural Science Foundation (Nos. 7232150, 7242029, 7242028 and 7244449) NSFC (Nos. 82370083, 82200091, 82341089, 82270279 and 82200296).
Author information
Authors and Affiliations
Contributions
Yingzhen Du, Jialei Li, Zhe Dai, Yao Zhao, Pingjun Zhu conceived the original experiments. Jialei Li, Yuxin Chen, Xiaoman Liu and Tian Xia contributed to the manuscript revision. Zhe Dai and Jialei Li carried out all the in vivo experiments and molecular investigation in vitro. Pingjun Zhu and Tian Xia wrote the whole manuscript. Pingjun Zhu contributed to study concept and design, obtained funding, supervision of the study, literature search, writing of the manuscript and revised the final version of manuscript. All the authors read the article and approved the submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Experiments were performed under a project license (Beijing, China) granted by the committee on the Ethics of Chinese PLA General Hospital, in compliance with the Guidelines for the Management and Use of Laboratory Animals.
Conflict of interest
The authors declared no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Du, Y., Li, J., Dai, Z. et al. Pyruvate kinase M2 sustains cardiac mitochondrial quality surveillance in septic cardiomyopathy by regulating prohibitin 2 abundance via S91 phosphorylation. Cell. Mol. Life Sci. 81, 254 (2024). https://doi.org/10.1007/s00018-024-05253-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05253-9